Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
First Close-up of Amyloid Fibrils
Steric zipper appears to be key structure in disease-associated protein aggregates
by Stu Borman
June 13, 2005
| A version of this story appeared in
Volume 83, Issue 24
PROTEIN STRUCTURE

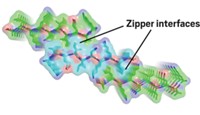
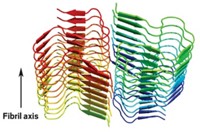
The first detailed atomic view of amyloid fibrils indicates that pairs of tightly interdigitating β-sheets are a common structural feature in these misfolded protein aggregates.
Amyloid fibrils are associated with some 20 diseases, ranging from Alzheimer's and type 2 diabetes to prion diseases. The new structure could aid the search for therapeutic agents that can impede formation or accelerate breakdown of amyloid in these conditions.
Amyloid fibrils formed from different proteins are known to have common structural properties, such as a "cross- βspine," a set of protein β-strands that run perpendicular to each fibril's long axis. An atomic model of such fibrils has not been available until now, however, primarily because of the difficulty in creating amyloid crystals large enough to be analyzed by X-ray crystallography.
"For more than 30 years, scientists have wondered what the molecular-level details are of the cross-β structure," says Howard Hughes Medical Institute investigator David Eisenberg of the University of California, Los Angeles, who led the group that carried out the new study (Nature 2005, 435, 773). "We have been fortunate to see the answer."
The work shows that a six- or seven-residue protein segment "can bind to other copies of itself to form two tightly interdigitating β-sheets," Eisenberg explains. "We call this tight interdigitation a 'steric zipper.' As the two sheets zip up, water is excluded, giving a dry, stable interface that is hard to reopen." The structure differs considerably from other known β-sheet architectures, and the zipper feature helps explain why amyloid fibrils are as stable and persistent as they are.
To solve the structure, Eisenberg and coworkers laboriously grew tiny crystals of amyloid fibrils--about 1/50,000th the size of conventional protein crystals, at best. They then used special techniques for structural analysis of very small crystals developed by crystallographer Christian Riekel and coworkers at the European Synchrotron Radiation Facility, Grenoble, France.
In a Nature commentary, professor of chemical and structural biology Christopher M. Dobson of Cambridge University calls the study "a breakthrough." And professor of cellular and molecular pharmacology Jonathan S. Weissman of the University of California, San Francisco, says determining the structure "is a monumental achievement that will open up a new era in the structural analysis of amyloids."



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter