Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Detecting Drugs' Influence
Technique reveals compounds' direct effects on protein signaling interactions
by Stu Borman
May 15, 2006
| A version of this story appeared in
Volume 84, Issue 20

A technique that detects the ability of compounds to affect cell-signaling pathways could aid drug research by making it easier to understand mechanisms and side effects of drug candidates.
The technique, PCA (protein-fragment complementation assay), was developed over the past decade by biochemistry professor Stephen Michnick of the University of Montreal and coworkers as a means for predicting unknown drug activities and toxicities. Michnick, in collaboration with John K. Westwick, president and chief scientific officer of San Ramon, Calif.-based Odyssey Thera, and coworkers have now demonstrated the technique on existing drugs (Nat. Chem. Biol., published online May 7, dx.doi.org/10.1038/nchembio790). Odyssey Thera, cofounded by Michnick, has an exclusive license for PCA and is developing a PCA-based drug screening program.
Robert T. Abraham, vice president of oncology discovery at Wyeth, comments that PCA could improve the efficiency and probability of success of drug R&D programs by making it easier to find new uses for existing agents and to screen out potentially toxic compounds at an early stage.
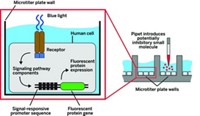
In PCA, two proteins that are known to interact in a biochemical pathway (such as a hormone and its receptor) are modified to include fragments of a fluorescent protein and then expressed in cells. If an introduced compound causes the two proteins to interact, the fragments combine, fluorescence is generated, and the signal is monitored microscopically, showing the cellular location and timing of the induced protein interaction. Measures are taken to distinguish signals induced by introduced compounds from those induced by endogenous cell components.
The paper describes the use of 49 different PCA tests to screen each of 107 known drugs. Test results were in line with the drugs' known structure-activity properties. The study uncovered previously unknown antiproliferative activities for four of the drugs, including the antidepressant sertraline.
Tim Mitchison, professor of systems biology at Harvard Medical School, comments that PCA might not be any better than current drug assessment techniques, such as gene expression array analysis, and might be more difficult to carry out. Gene expression analysis measures effects of bioactive compounds on protein production in cells. Michnick, Westwick, and coworkers "have identified a real problem and made some progress on solving it, but I doubt it's the be-all and end-all" of drug effects analysis, Mitchison says.
Michnick replies that PCA captures responses at steps of single biochemical pathways, whereas gene expression changes are influenced by multiple pathways, making it difficult to attribute drug effects to specific targets. "I hope this report will inspire others to use PCA, develop other approaches for direct pathway detection, and think more about how to interpret such experiments," he says.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter