Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Complexity To Live By
This year's Nobel Prize In Chemistry highlights the elegant complexity biology musters to read genes
by Ivan Amato
November 20, 2006
| A version of this story appeared in
Volume 84, Issue 47

Every year, a few amateur mushroom collectors discover what happens when the molecular machinery at the center of this year's Nobel Prize in Chemistry, the RNA polymerase II (pol II) complex, gets poisoned.
When people eat Amanita phalloides mushrooms, their digestive systems begin extracting the toxin α-amanitin, which travels through the bloodstream to every cell in their bodies. As the hours and days pass, molecules of this bicyclic octapeptide find their way into the cells' nuclei and nestle into the pockets of countless copies of the RNA pol II enzyme.
When unfettered by α-amanitin, (pol II) transcribes protein-encoding stretches of nuclear DNA—genes, that is—into corresponding stretches of messenger RNA. The resulting strands of mRNA leave the nucleus and are translated into protein by another amazingly complex molecular machine, the ribosome. It was partly for his atomic-scale portraiture of pol II, this astounding DNA-to-RNA transcription enzyme, that Roger Kornberg of Stanford University received the coveted call from Stockholm last month.
If all of a person's pol II were to suddenly disappear or get poisoned into inaction, all of the genes in every cell of the person's body would instantly become analogous to an archived library that never could be accessed again. Genes would be locked away. All protein synthesis would come to a halt. Life would peter out, as it does each year for a small club of unfortunate mushroom adventurers. The common name for A. phalloides is the Death Cap mushroom.
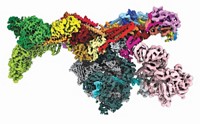
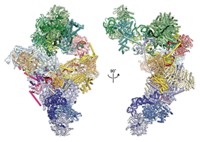
On Oct. 4, in a phone call from Sweden at 2:30 AM PST, Kornberg learned that he was this year's winner of the Nobel Prize in Chemistry for his efforts to shed light on the molecular architecture of RNA pol II and how it transcribes DNA into RNA. Twenty years in the making, and relying on a huge number of fragile, hard-won crystals analyzed with some of the brightest X-ray sources in the world, the achievements most emblematic of the award were published in a pair of papers that appeared in 2001 (Science 2001, 292, 1863 and 1876). The first paper describes yeast's 10-subunit pol II at a resolution of 2.8 Å, fine enough for Kornberg and his coworkers to begin to see the atomic-scale mechanical details. The second paper provides a crystal snapshot at 3.3-Å resolution of pol II in the act of transcribing DNA into an mRNA molecule.
The two papers are rife with mechanical language, describing the components of the polymerase with words like hinge, wall, loop, clamp, funnel, and jaw. Now, when Kornberg's group and other researchers talk about transcription by the polymerase, they can refer to specific chemical locations where DNA attaches, where and how the double-stranded DNA is pried apart, where and how the template strand of DNA is fed into the enzyme's active site, where and how the corresponding RNA building blocks are delivered and then chemically stitched into mRNA, and how the nascent mRNA strand unwinds from the enzyme while the temporarily separated strands of DNA zip back together into a double helix.
"At its core, it is a chemical reaction," David Bushnell reminded millions of listeners in an Oct. 4 interview on National Public Radio. Bushnell, a senior scientist now in his 13th year with the Kornberg lab, pointed out that their structures have revealed how the enzyme catalyzes the formation of phosphodiester bonds between nucleotides of a growing RNA strand. Bushnell and labmate Patrick Cramer, now at the University of Munich, Germany, screened an estimated 1,000 crystals over the years to produce the kinds of crystallographic data required for this level of chemical understanding.
"We have only 20% of the machine at this level of detail," says Kornberg, as a reminder of how much work still needed to be done. "We want the whole machine."
In all, researchers have identified some 60 protein subunits as components of the transcription system. Among these is a massive, multicomponent unit that Kornberg's group identified and dubbed Mediator in the 1990s because it helps integrate inputs from a plethora of gene-controlling factors that influence transcription.
Kornberg already has received numerous awards for his work on transcription. Talk of the Nobel caliber of that work, as well as of earlier work on the structure of chromosomes, has been perennial for some time. Last month's Nobel announcement has come with widespread applause.
"Roger is richly deserving of this recognition," comments Harry Noller of the University of California, San Francisco. Noller, who investigates the structure and function of the ribosome, where RNA is translated into protein, adds that Kornberg's "work has provided the structural basis for understanding how RNA is polymerized from a DNA template."
Of course, asking and answering gargantuan scientific questions—such as, "How does the molecular machinery by which genes are translated into mRNA work?"—is never owned by a single scientist. Although everyone whom C&EN contacted agrees that Kornberg was fully deserving of a Nobel Prize, some also shared their surprise that he was the sole winner.
In an e-mail exchange, for example, Phillip A. Sharp of Massachusetts Institute of Technology praises Kornberg's achievements. "The atomic structure of the RNA pol II complex determined by Kornberg is a singular advance," he writes. But Sharp, who shared the 1993 Nobel Prize in Physiology or Medicine, also expressed some puzzlement: "The question of this prize is how they could have not included Robert G. Roeder at Rockefeller University, who was among the first to identify RNA pol II." Sharp adds that Roeder led "the characterization of the activities of the polymerase in regulation of genes" for decades.
In the late 1960s, Roeder identified three RNA polymerases, designated pol I, pol II, and pol III, with distinct enzymatic properties and subcellular localizations. With a colleague, he used the Death Cap mushroom toxin to show that these enzymes had distinct functions. He found that the different polymerases were affected by the toxin to greatly different degrees. He went on to show that pol I orchestrates the production of RNA earmarked for ribosome construction, that pol II produces the mRNA necessary for gene transcription, and that pol III makes the transfer RNA molecules that deliver amino acids to ribosomes during the construction of proteins.
Roeder and colleagues also contributed to the identification and characterization of many so-called transcription factors and other components without which pol II cannot work properly. These include a convoy of transcription factors that snap together in sequence at specific chromosomal locations and then, once assembled, recruit pol II to the site. Because of these and many other discoveries over the decades, Sharp and other observers think that Roeder in particular, among the many researchers who have been contributing to the transcription story, could have shared the prize with Kornberg.
Roeder, who has won his share of awards, including the coveted Lasker Award in 2003, told C&EN that he is "a little bit disappointed by this whole thing," but he also is not dwelling on missing the golden ring this time around. Kornberg has been doing "spectacular structural work" he says, and this year's award cites just that aspect of the Nobel-winning work. "Roger Kornberg was the first to create an actual picture of how transcription works at the molecular level in the important group of organisms called eukaryotes," states the Royal Swedish Academy of Sciences in the press release announcing the award. Eukaryotes, including all mammals, are organisms whose cells have membrane-enclosed nuclei, as opposed to prokaryotes, such as bacteria, which do not have nuclei. Prokaryotes have an evolutionarily earlier and simpler version of the eukaryotic machinery that Kornberg has focused on.
As Kornberg sees it, the solo nature of the chemistry award reflects a vast research enterprise involving many talents of the highest order. "What I say to people is that the Nobel Committee for Chemistry singled out something that is a culmination of everything," he says.
One of the happiest realizations in the earlier days of his work on transcription, Kornberg says, was that the molecular machinery of transcription is virtually identical in all eukaryotic cells, whether they be of yeast, fruit, or Nobel Laureate. This yeast-to-beast similitude gave the researchers confidence that their experimentally convenient choice of studying the transcription mechanism in the microorganism that makes bread rise would yield knowledge directly relevant to the way human cells live, replicate, differentiate, and organize into a living, breathing person. In the long term, researchers say, understanding gene transcription and how it is controlled by pol II has bearing on everything biological and many things medical, including possibilities for tweaking cancer cells back to healthy genetic settings and steering stem cells to become specific cell types.
Trained as a chemist at Harvard University and Stanford University, Kornberg, 59, always has had a leaning toward, in his words, "life chemistry." His family may have something to do with this. When Roger was a teenager, his father, Arthur Kornberg, was elucidating the mechanisms by which cells synthesize DNA, work for which the senior Kornberg shared the 1959 Nobel Prize in Physiology or Medicine. Roger's mother, the late Sylvy Kornberg, was a biochemist. One of his brothers, Thomas Kornberg, investigates the molecular biology and genetics of development in his lab at UC San Francisco.
In his own graduate work in the 1960s, Kornberg focused on the chemistry of a very different part of cellular anatomy: the lipid bilayer membrane, which determines what goes in and out of a cell. Working with longtime Stanford physical chemist Harden McConnell, Kornberg developed a feel for the ways molecules move through and within membranes. He did not know it at the time, but he and coworkers later would tap into this work to reveal pol II's detailed structure. In particular, they found that lipid membranes could be used to organize hard-to-crystallize pol II into 2-D crystals. They eventually used these as seeds to produce the 3-D crystals they needed to achieve the atomic-level resolution they rolled out in their 2001 Science papers.
Always adamant that a precise knowledge of structure was central to understanding biology from a chemical perspective, Kornberg secured a postdoctoral position in the early 1970s at the Medical Research Council's (MRC) Laboratory of Molecular Biology in Cambridge, England. There he learned electron and X-ray crystallography techniques from Aaron Klug, who would win a Nobel Prize in Chemistry in 1982. There Kornberg also developed a fascination with chromosome structure-the overall macromolecular context in which eukaryotic transcription occurs-from none other than Francis Crick, who shared a Nobel Prize in 1962 for uncovering the structure of DNA.
Kornberg decided to apply the structure elucidation methods he was learning at MRC to chromatin. The stuff of chromosomes, chromatin was known to be composed of DNA and proteins known as histones, but its precise molecular organization remained mysterious. In August 1973, Kornberg made what turned out to be a correct and revelatory conjecture about chromatin structure. As he imagined it, octets of histone proteins organized into thousands upon thousands of spoollike structures around which DNA strands wound two times. Strung along the chromosome-like beads, these so-called nucleosomes, which also interact to form higher levels of chromosomal organization, provide the structural means by which a meter or so of DNA manages to jam into a nucleus only a few micrometers in diameter.
As usual, a scientific insight of this magnitude raised equally vast questions. How could such a tightly scrunched chromatin structure support the selective transcription of genes distributed here and there throughout a chromosome's length? The transcription of genes at specific times and under particular biochemical circumstances became compellingly mysterious in the new chromatin context that was emerging. To Kornberg, the answer in chemical terms would come only when the structures of the transcription machinery were revealed in sufficient detail.
To get this picture for pol II, Kornberg and his coworkers had to find ways of crystallizing the enzyme and various combinations of the many transcription factors and other protein components that assemble into what Sharp describes as "the largest and most complex nonrepetitive protein structure reported to date."
Advertisement
Procuring pol II took dogged devotion by Kornberg, postdocs, graduate students, and other collaborators. "We could only get about 25 µg from 1 L of yeast culture, but we needed hundreds of milligrams to make a crystal," Kornberg says. Throughout the 1980s, the researchers went through 10,000 L of yeast culture before they were able to produce their first 3-D crystal of pol II in 1990.
Key to their work, Kornberg and Bushnell stress, was that the lab is a short 10- or 45-minute drive, respectively, from synchrotron X-ray sources at the Stanford Linear Accelerator Center (SLAC) and the Advanced Light Source at Lawrence Berkeley National Laboratory. X-ray scattering from the crystals produces diffraction patterns from which researchers can reconstruct the crystals' specific arrangement of atoms.
"I can't tell you how many sleepless nights we've had" during these crystallography runs, Bushnell says, adding an observation that reflects how Kornberg has run his laboratory. "He used to show up after the kids went to bed to sit there and be there at the synchrotron with us," Bushnell says.
On Dec. 8, Kornberg will deliver his Nobel Lecture in Chemistry at the Aula Magna, Stockholm University. From there, he'll continue going after ever finer views of the machinery of life.
Read More
- 100 Years Ago
- 1906 Chemistry Nobelist Henri Moissan Spawned The Vast Arena Of Fluorine Chemistry






Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter