Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
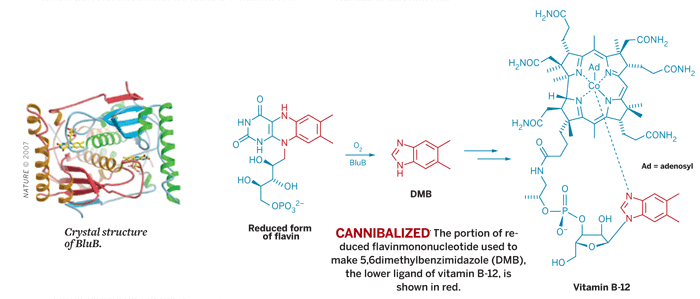
Vitamin B-12 Mystery Is Solved
Enzyme structure reveals that one vitamin is cannibalized to make another
by Celia Henry Arnaud
March 26, 2007
| A version of this story appeared in
Volume 85, Issue 13
BY CAPTURING the structure of the sole missing link in its biosynthesis, scientists have solved a long-standing mystery surrounding vitamin B-12.
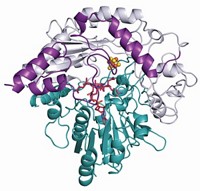
Vitamin B-12 is one of the largest of the known nonpolymeric natural products. Its biosynthesis is well-understood, with one notable exception: Scientists have struggled to figure out how the vitamin's 5,6-dimethylbenzimidazole (DMB) ligand is made. Biologist Graham C. Walker of MIT, biochemist Christopher T. Walsh and crystallographer Nicholas A. Larsen of Harvard Medical School, and coworkers now have filled in that knowledge gap by solving the crystal structure of BluB, the enzyme that synthesizes DMB (Nature 2007, 446, 449).
They find that the enzyme cannibalizes one vitamin to make another. BluB forms DMB by fragmenting and contracting flavin mononucleotide, a cofactor derived from vitamin B-2. In the structure they have determined, the researchers captured an early BluB intermediate in which molecular oxygen is poised to attack the flavin.
Flavin as a substrate is unusual. It more commonly acts as a cofactor in reactions rather than being itself metabolized. "Part of what is so exciting about this is that something that is normally a cofactor is getting cannibalized and made into part of another cofactor," Walker says.
On the basis of the enzyme's amino acid sequence, the researchers initially thought that it might be a member of a family of enzymes that use the cofactors NADH or NADPH (the reduced forms of nicotinamide adenine dinucleotide and nicotinamide adenine dinucleotide phosphate, respectively) to reduce flavin for a variety of reactions. Kinetic data quickly dispelled the notion that BluB acts similarly, because the concentrations of NADH and NADPH that it requires to reduce flavin are well above physiological concentrations. Instead, the researchers suggest that the enzyme represents a new family of "flavin destructase" enzymes.
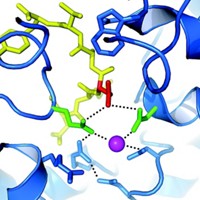
BluB's active site turns out to be surprisingly small. Before the scientists analyzed the kinetic data, they thought that the enzyme would bind NADH to reduce the flavin. "We thought that we would see a nice big pocket with room for NADH to go in," Larsen says. Instead, the active site is a tight squeeze for already reduced flavin and molecular oxygen. On one side of the active site, a helix also found in another family of enzymes known as oxidoreductases is lengthened so that it acts as a lid that completely buries the flavin. Other helices are shorter or missing, further tightening BluB's binding pocket.
"One has to marvel at how such a small enzyme is able to perform this catalysis, which involves the breaking and making of so many carbon-carbon bonds, including the surgical excision of a carbon from a six-membered ring to generate a five-membered ring," says Martin J. Warren, a professor of biochemistry at the University of Kent, in Canterbury, England.
The researchers don't yet know the details of the mechanism by which BluB transforms flavin into DMB. Two amino acid residues—an aspartic acid and a serine—appear to be key. Mutating the serine to glycine reduces DMB production 30-fold, and mutating it to cysteine wipes out DMB production. Mutating the aspartic acid still allows the enzyme to bind flavin but eliminates DMB production, a result suggesting that aspartic acid is essential for catalysis.
In an accompanying commentary, chemists Steven E. Ealick and Tadhg P. Begley of Cornell University suggest a possible mechanism "in which a series of reactions with water dismantles the flavin after it has reacted with molecular oxygen" (Nature 2007, 446, 387).


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter