Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Flu Virus Proton Channel Analyzed
Structures of key surface protein suggest different drug mechanisms
by Stu Borman
February 11, 2008
| A version of this story appeared in
Volume 86, Issue 6
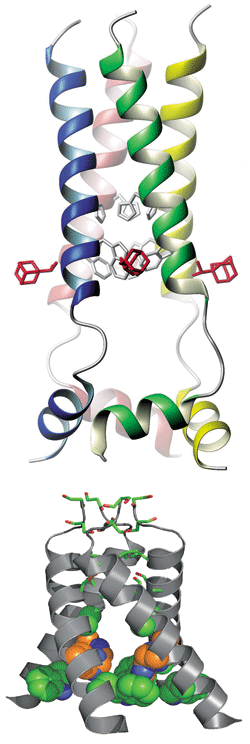
NMR structure of the closed state of M2 (top, with three bound drug molecules visible in red) and X-ray crystal structure of the channel protein's open state (bottom, not including bound drug).
TWO RESEARCH GROUPS have independently used different methods to obtain long-sought structures of M2, a key ion-channel protein on the surface of influenza virus. The work could help scientists figure out why the virus has recently become resistant to two of the four drugs approved as antiflu agents. It could also lead to the design of more-effective medications.
But scientists will first have to better understand the implications of the new findings. The two studies agree in some ways about the architecture of the flu virus ion channel but come to opposite conclusions about where the drugs bind—whether to the channel's open or closed state—and the mechanism by which they block the channel's activity.
Only two antiflu drugs—Tamiflu (oseltamivir) and Relenza (zanamivir)—are currently recommended in the U.S. to help alleviate flu symptoms. Two other drugs—Symmetrel (amantadine) and Flumadine (rimantadine)—are approved agents, but they are no longer recommended because most strains of flu virus have become resistant to them in the past few years. The World Health Organization reported last month that a Tamiflu-resistant strain is becoming more prevalent.
A few years ago, M2 mutations caused most strains of flu virus type A to become amantadine- and rimantadine-resistant, perhaps in response to overuse of the drugs. A second class of virus, type B, also causes flu but has a different proton-channel surface protein, called BM2, which isn't inhibited by any of the four approved antiflu drugs.
To find out how and why flu virus has learned to so effectively evade amantadine and rimantadine, scientists have been seeking the structure of their molecular target, the ion-channel protein M2 on the flu virus surface. Tamiflu and Relenza have different targets.
M2 has proved difficult to analyze, but two teams have now succeeded: structural biologist James J. Chou and postdoc Jason R. Schnell of Harvard Medical School, using nuclear magnetic resonance spectroscopy (NMR); and macromolecular structure and function specialist William F. DeGrado of the University of Pennsylvania and coworkers, using X-ray crystallography (Nature 2008, 451, 591 and 596).
M2 is a membrane-spanning ion channel that conducts only protons and is required for viral infectivity. With only 97 amino acids in each of its four identical peptide subunits, it's considered to be a minimal channel protein. This is the first proton channel to have been analyzed structurally, so the studies should help researchers learn more about the way such channels work.
The Harvard structure shows the channel protein in a closed (ion-nonconductive) state with the drug molecule bound on the outside of the channel, near the channel's cytoplasmic end. The Penn picture shows an open channel with one drug molecule plugging the pore interior, near the channel's extracellular end. The two structures suggest different molecular mechanisms of drug action.
THE FORMS OF M2 analyzed in the two studies are merely small fragments of the tiny protein. The Chou and DeGrado groups each analyzed segments of only one of the tetrameric protein's four peptides. The crystallographic study was carried out on the peptide's 25-residue membrane-spanning part, and the NMR structure was obtained on a peptide comprising the membrane-spanning segment plus 18 additional amino acids.
The two structures show M2 to be a conical membrane-spanning channel protein, with a narrow constriction on its extracellular end and a wider opening on its cytoplasmic end. The Harvard structure of the closed channel is less conical than the Penn structure of the open form. The wide part of the cone has a tryptophan "gate," which opens or closes the channel in response to pH changes, and a histidine that serves as a pH sensor. The channel is open when the extracellular medium is acidic (around pH 6) and closed when it's at neutral pH.
Amantadine and rimantadine block the native, unmutated M2 channel, making the virus noninfective. The drug molecules unexpectedly turned up in different locations in each structure. The results lead to different predictions about how the drugs might turn off the channel and how resistance mutations might disable the drugs.
In the DeGrado group's open-channel structure, one molecule of drug seems to bind inside the pore near its constricted end and to work by plugging it directly. The Chou group's closed-channel structure indicates that one drug molecule per subunit (up to four per channel) binds to the exterior of the channel and that the drug shuts down the channel by stabilizing the closed state thermodynamically.
The Penn and Harvard groups "are dramatically disagreeing mechanistically," Christopher Miller tells C&EN. Miller, a Brandeis University professor of biochemistry who specializes in ion-channel structure and function, also wrote a commentary on the studies (Nature 2008, 451, 532). "The idea is that you do a structure not just to get a pretty picture of a protein, but so you can understand how it works. The messages from these structures about how the drugs work are completely contradictory."
The direct blockage mechanism proposed by the Penn group is feasible because some key M2 mutations surround the crystallographically defined drug-binding site, where they are well-positioned to make it difficult for the drug to bind. The Harvard group's mechanism is also feasible because resistance mutations might destabilize the closed state and thus promote the open state, countering the inhibitory effect of the bound drug.
"Each of those pictures gives its own biophysical predictions for how the channel should behave with drugs at different pH values and so on," Miller says. "These are things that can be tested" so issues about binding and mechanism can be resolved.
Professor of neurobiology and physiology Lawrence H. Pinto of Northwestern University, who codiscovered M2's and BM2's roles as ion channels, comments that "the two papers serve as a signal that people interested in developing the next generation of M2 inhibitors should get to work. There is enough information in the papers to tackle the problem of making useful inhibitors." Pinto says he hopes the studies "also stir up interest in developing inhibitors for the BM2 channel of influenza type-B virus, which is responsible for about half of the flu morbidity in a typical year."
Researchers in the field "all agree on the need to do more experiments, both structural and biochemical, to nail down the mechanism" of M2 inhibitors, Chou says. "We are working intensely on follow-ups."
DeGrado believes the new structures "will spur a variety of experimental studies to resolve the mechanism of inhibition. Ultimately, these studies should lead to new, better inhibitors for endemic outbreaks of influenza as well as future pandemics."


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter