Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Enzyme Makes A Tough Cut
Crystal structure of bacterial enzyme suggests route to a challenging bond cleavage
by Carmen Drahl
June 15, 2009
| A version of this story appeared in
Volume 87, Issue 24

With the help of X-ray crystallography, researchers have identified and structurally analyzed an iron-containing enzyme that carries out a tricky bond-breaking reaction. The findings could inspire versatile new catalysts.
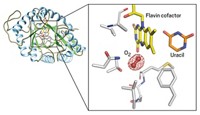
Bacteria make phosphinothricin, a natural product widely used as a weed killer, with several unusual steps. One includes the cleavage of a very poorly activated carbon-carbon bond in 2-hydroxyethylphosphonate (HEP), a phosphinothricin precursor.
Because unactivated C–C bonds are tough to break with the known synthetic tool kit, scientists want to understand how this transformation takes place, says chemist Wilfred A. van der Donk of the University of Illinois, Urbana-Champaign, who has been working to understand phosphinothricin biosynthesis. But until now his team "didn't know what kind of enzyme did the job and whether cofactors were required for it to work," he says.
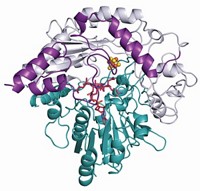
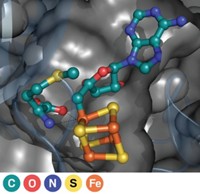
So van der Donk's group joined forces with structural biologist Satish K. Nair and microbiologist William W. Metcalf, both at Illinois. The team has now solved the enzyme's X-ray structure and identified it as a nonheme iron-containing oxygenase (Nature, DOI: 10.1038/nature07972). The enzyme, hydroxyethylphosphonate dioxygenase (HEPD), uses molecular oxygen to cleave the poorly activated C–C bond in HEP.
The team found that HEPD doesn't need cofactors to do its job, unlike most other dioxygenase enzymes. Instead, they speculate that HEPD begins its catalytic cycle with iron in the +2 oxidation state and that the enzyme reacts with oxygen to form an Fe(III) superoxide species, which initiates the bond cleavage by plucking a hydrogen atom from HEP.
That superoxide species has been described in other iron enzymes, including one that cleaves C–C bonds, say J. Martin Bollinger Jr. and Carsten Krebs, who run a lab studying metalloenzymes at Pennsylvania State University. The Illinois team hasn't observed the Fe(III) superoxide directly, but a combination of techniques including Mössbauer spectroscopy might confirm its role in the bond-cleaving mechanism, they say. "By learning about these enzymes, we hope new chemical or catalytic processes will emerge," they add.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter