Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Ubiquitin Tagging On Demand
by Carmen Drahl
November 16, 2009
| A version of this story appeared in
Volume 87, Issue 46
Having the right tools is the key to understanding ubiquitination, or the tagging of proteins with ubiquitin that marks them for disposal. Thanks to chemical biologists, one of those tools, the ubiquitin-tagged protein, is getting easier to come by. Ubiquitin-tagged proteins are essential for studying structures of proteins in the pathway, dissecting signaling networks, and more.
Making a ubiquitinated protein is all about forging a ubiquitin isopeptide bond. Unlike regular peptide bonds, these amide bonds link the amine on the tip of a lysine side chain of the protein to the carboxyl group on ubiquitin's C-terminal glycine.
Traditionally, researchers make ubiquitin-tagged proteins with E1, E2, and E3, the enzymes in the ubiquitin-proteasome pathway that mark a protein for destruction. It is a complicated proposition, and one that isn't always possible because researchers haven't yet identified the tagging machinery that goes with each and every protein.
On top of that, unlike most other protein-tagging systems, such as phosphorylation, ubiquitination can result in attachment of one ubiquitin or any number of ubiquitins to a protein. It's difficult to control how many ubiquitins end up on a protein, and the resulting heterogeneous samples aren't useful for dissecting the roles of a given ubiquitination pattern in the cell, something that's still largely a mystery.
Several recently developed techniques now make it easier to produce pure samples of ubiquitin-tagged proteins specifically labeled at a desired lysine side chain. A selection of the approaches uses native chemical ligation (NCL), an established method for protein chemical synthesis. NCL requires a 1-amino, 2-thiol moiety, traditionally from a cysteine residue, on one ligation partner and a thioester moiety on the other.
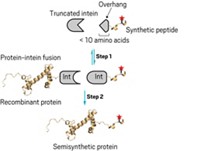
To use NCL for ubiquitination, a team led by Rockefeller University chemist Tom W. Muir developed a synthetic linker containing two orthogonal amino-thiol handles. Each directs formation of an amide bond with a thioester group, one on ubiquitin, the other on a protein of interest. Subsequent removal of the sulfur-containing handles leaves behind a ubiquitinated protein (Nature 2008, 453, 812). Muir's team recently developed a more efficient version of this technique that eliminates a bulky intermediate (ACS. Chem. Biol., DOI: 10.1021/cb9002255).
Prelude To A Kiss Of Death
Targeting early steps in protein disposal could lead to drugs for cancer and more
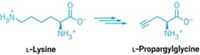
Meanwhile, Ashraf Brik and coworkers at Ben Gurion University of the Negev, in Beer Sheva, Israel, put a thiol group directly on the δ carbon of a lysine side chain. They incorporated the modified lysine into a 17-residue peptide derived from the protein α-synuclein and demonstrated that a temporary thiol handle at this location could facilitate ubiquitination (Angew. Chem. Int. Ed. 2009, 48, 8090).
By moving the thiol handle just one carbon down the lysine side chain, to the γ position, researchers in Singapore could direct two ligations for the price of one (J. Am. Chem. Soc. 2009, 131, 13592). Using their γ-thiol modified lysine, Xue-Wei Liu, Chuan-Fa Liu, and coworkers at Nanyang Technological University successfully placed a ubiquitin tag on a 12-amino-acid peptide (C&EN, Sept. 21, page 35).
Most recently, Michael K. Chan and coworkers at Ohio State University have taken a more biological approach to site-specific ubiquitination. Their ligation relies on a cysteine-containing analog of pyrrolysine, the 22nd genetically encoded amino acid. Chan previously discovered pyrrolysine together with Ohio State colleague Joseph A. Krzycki. With the help of the biochemical machinery in Escherichia coli bacteria, Chan and coworkers can genetically encode pyrrolysine into any protein they desire. Now, they've done the same for a new 1-amino-, 2-thiol-containing pyrrolysine analog (Angew. Chem., DOI: 10.1002/anie.200904472). Using that method and combining it with NCL, the researchers believe they can attach ubiquitin to virtually any protein.
The four sets of work "make ingenious use of native chemical ligation to achieve site-specific ubiquitination of proteins," says Stephen B. H. Kent, a pioneer in protein chemical synthesis at the University of Chicago. "They are complementary yet distinct in their respective approaches. This should be of considerable interest to the chemical biology research community."



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter