Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Shaping Up Protein NMR
Spectroscopy: Techniques borrowed from solid-state NMR sharpen proteins' broadened spectral lines
by Carmen Drahl
June 30, 2010
| A version of this story appeared in
Volume 88, Issue 27

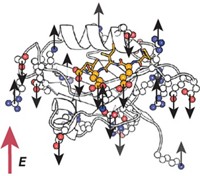
By adapting two techniques from solid-state nuclear magnetic resonance, chemists have found a new way to enhance resolution in protein NMR (J. Am. Chem. Soc., DOI: 10.1021/ja103251h). The method could provide a sharper view of proteins in motion than is now possible with NMR.
Researchers studying protein dynamics by NMR often run into a vexing problem: The very events they wish to study, such as protein folding or ligand binding, can broaden resonances in NMR spectra, sometimes so much that it's hard to extract useful information.
"What one would like to be able to do is narrow the lines when recording the protein NMR spectrum," to get the best resolution and sensitivity possible, says Arthur G. Palmer III of Columbia University. "Then when capturing conformational dynamics," where broadened resonances are useful, "one could turn the broadening back on," he says.
Now, Palmer and postdoctoral researcher Ying Li have done just that. They borrowed a pair of homonuclear dipolar decoupling methods from solid-state NMR and demonstrated the narrowing effect with the protein RNAse A. The technique, which they plan to extend to more proteins, can be combined with other resolution enhancement methods, Palmer notes.
"It will be exciting to see how this method helps in some biologically significant and previously intractable samples," says Lila M. Gierasch, an expert in protein NMR at the University of Massachusetts, Amherst. "Biological function requires dynamics, and this method promises to open the door to systems that refuse to sit still."


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter