Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Putting DNA In A Bind
ACS Meeting News: Small molecules that interact with DNA are bound to regulate gene expression
by Stu Borman
September 27, 2010
| A version of this story appeared in
Volume 88, Issue 39

A number of important drugs do their work in the body by binding to double-stranded (duplex) DNA. Inspired by the success of these drugs, scientists are trying to design other synthetic agents that work the same way.
The rationale for these efforts is that “you want to be able to regulate gene expression,” according to molecular recognition specialist Dev P. Arya of Clemson University. “If you can find agents that target specific DNA sequences and structures, you can use them to target specific diseases or as molecular biology tools to study DNA.”
One drug that targets duplex DNA is cisplatin, a platinum-based anticancer agent that binds to guanine residues in the major groove of DNA. And anthracycline anticancer agents such as daunomycin and adriamycin have aromatic rings that intercalate between base pairs of DNA as well as extra groups that then bind to DNA’s minor or major groove. But current DNA-duplex-binding drugs “tend to be rather toxic,” Arya told C&EN. “So approaches that allow better recognition of DNA will lead to better drugs, as well as better probes of DNA biology down the line.”
Many human diseases have been traced to problems with the expression of specific genes, and to fix such problems scientists need to figure out how “to do DNA-sequence-specific recognition,” according to Larry W. McLaughlin of Boston College, who specializes in targeting duplex DNA. “If you could do that, it would open up whole new classes of antiviral or anticancer compounds, where you could go in and specifically target gene promoters and shut them down.”
To discuss how to recognize DNA in a sequence-specific way, Arya, McLaughlin, and others joined together at a Division of Carbohydrate Chemistry symposium titled “Recognition of DNA: Recent Advances” at last month’s American Chemical Society national meeting in Boston. Arya organized and presided over the session, which covered a number of strategies for recognizing duplex DNA.
Presentations during the session included discussions of the traditional way of targeting DNA duplexes—designing small molecules that target the minor groove, one of the two key structural features of the helical macromolecule. For reasons not yet completely understood, scientists have had far less luck finding small molecules that bind to DNA’s major groove, its other major structural motif. Arya described how his group identified the first synthetic molecules that do so. Researchers also discussed other nontraditional DNA recognition strategies. For example, McLaughlin and coworkers are developing a unique breed of triple-helix-forming binders. And there is a new emphasis in the field on targeting not just specific DNA sequences but also DNA sites having specific physical properties and shapes.
Inspired in part by the minor-groove-binding proclivities of the anticancer natural product distamycin, Peter B. Dervan of California Institute of Technology and colleagues have pioneered the development of polyamides as small-molecule DNA binders over the past few decades. Distamycin binds only to adenine-thymine (AT) base pairs, but Dervan and coworkers have used chemical synthesis to design polyamides that are sequence-specific for all four base-pair permutations—AT, TA, guanine-cytosine (GC), and CG.
Polyamides permeate cell membranes, bind to the minor groove of cellular DNA, and disrupt interactions between DNA and important transcription factors in the major groove. But it was never understood mechanistically how or why minor-groove binding could affect major-groove events. Dervan and grad student David M. Chenoweth, now an assistant professor of chemistry at the University of Pennsylvania, have answered that question by obtaining a high-resolution crystal structure of a U-shaped polyamide bound to DNA (J. Am. Chem. Soc., DOI: 10.1021/ja105068b).
The structure shows that polyamide binding widens the minor groove of DNA by 4 Å and, in an allosteric manner, compresses the adjacent major groove by 4 Å. This reveals why polyamides can exert such profound effects in the context of the cell. By binding to and modifying DNA’s minor groove, they change the structure of DNA’s major groove so a transcription factor can no longer bind there, thus inhibiting the expression of genes controlled by that interaction.
Also interested in minor-groove recognition are biophysical chemist W. David Wilson of Georgia State University and coworkers, who focus on small heterocyclic dications instead of polyamides as DNA-binding agents. “We’ve identified some clinically useful compounds against eukaryotic parasitic microorganisms that cause sleeping sickness, leishmaniasis, malaria, and other conditions,” Wilson said. “We’re trying to understand the fundamental principles of DNA recognition of these types of compounds to better understand what kind of biology they induce once they’re bound to DNA.”
A heterocyclic dication called furamidine, synthesized by Georgia State emeritus organic chemist David W. Boykin, showed excellent activity against sleeping sickness and went all the way to Phase III clinical trials in Africa in the form of an oral prodrug. But ultimately, the drug failed because of toxicity problems revealed in a follow-up Phase I trial. “The toxicity that appeared for a few people was very serious, so we couldn’t play around,” Wilson said. The failure was nevertheless a big disappointment.
Furamidine binds strongly to strings of AT base-pair sequences in DNA’s minor groove. “This is the key to selective targeting using small molecules—synergistic effects, where you have repeat sequences,” Wilson said. “The small molecules line up on a piece of DNA, and we think that’s necessary to do biology.”
Unlike Dervan and coworkers, Wilson said, “we don’t yet have compounds that can target different sequences. That’s one of the features we’re working toward.”
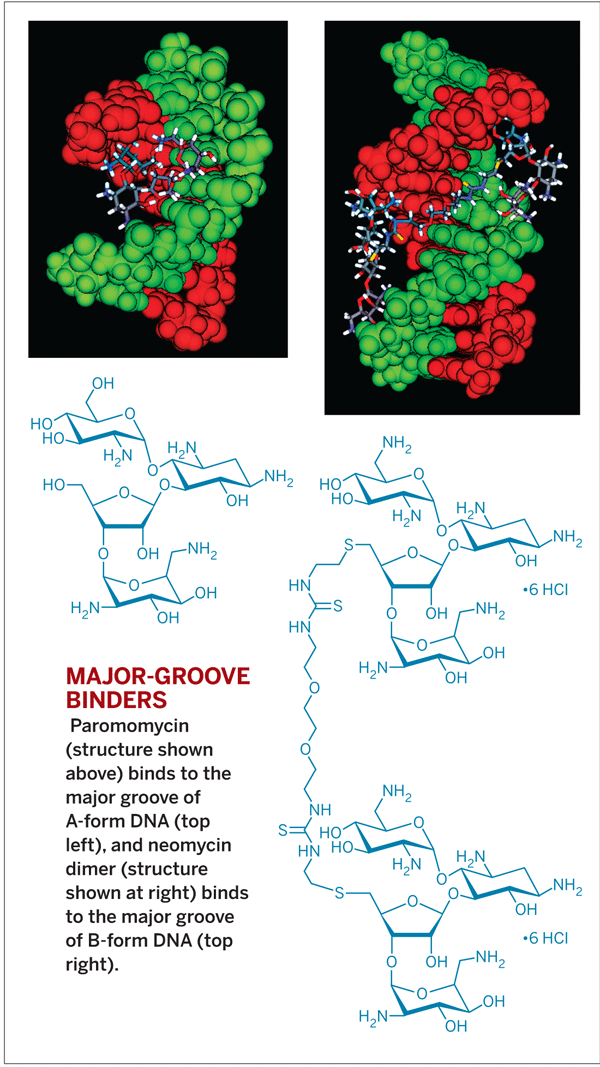
Although scientists could recognize the minor groove of DNA with polyamides or heterocyclic dications, they were initially out of luck targeting the major groove with small molecules. Arya and coworkers filled this DNA-recognition gap in 2003, when they found aminoglycosides such as paromomycin that recognize the major groove of A-form DNA. A- and B-form DNA are right-handed helical conformations of duplex DNA that predominate in nature; left-handed Z-form DNA is much rarer.
After finding A-form binders, Arya and coworkers wanted to identify B-form-targeted agents as well. The major groove of B-form DNA is wider than that of A-form, so they speculated that bigger small molecules would be needed to recognize it. “We did a lot of modeling and preliminary binding studies and hit upon dimeric aminoglycosides such as neomycin dimer that bind with high affinity in the major groove of B-form DNA,” Arya said.
The group now has lead molecules that each target both A- and B-form DNA. Their goal, like that of many others in the field, is to develop structure- or sequence-specific binders as novel DNA-targeted drugs.
Meanwhile, McLaughlin and coworkers have been working on a DNA-binding system that is unlike any other. In their unique system, a peptide nucleic acid—PNA, a nucleic acid analog—attaches to duplex DNA to form a “Janus wedge triplex.” Named after the Roman god with two faces, Janus wedge triplexes are formed by PNAs with residues that have two-faced hydrogen-bonding surfaces. Each residue wedges itself between the two bases of a base pair in the DNA duplex. The wedge residue thus takes custody of the hydrogen bonds that those two bases would normally share with one another.
“It is not really triplex formation in the minor or major groove exclusively,” McLaughlin said. “Here you have actually invaded the base pair, as opposed to just having bound on the outside of the base pair,” as in other groups’ triplex-forming systems. “Since the PNA invades the duplex, the Janus wedge residue has exposure from both the minor and major grooves.”
The overall goal of the group’s Janus wedge studies is to enable the agents to recognize all four DNA base pairs and thereby inhibit transcription at genes of interest for drug discovery applications.
A key trend in recent research on DNA-duplex recognition is a growing appreciation for the way DNA sequence affects localized physical properties and shape in the DNA helix. Pharmaceutical scientist Barry Gold of the University of Pittsburgh and coworkers have found that the electrostatic potential of specific sites on duplex DNA is sequence-dependent and that variations in electrostatic potential have a major influence on thermodynamic stability, ion binding, hydration, chemical reactivity, and protein interactions at the sites where they occur.
For instance, electrostatic potential changes can cause localized DNA instability, which is characterized by “a significant decrease in the water molecules associated with the grooves of DNA and also with the cations found in the grooves,” Gold said. “Such local instability may provide a mechanism for the recognition of DNA damage by DNA repair enzymes that very rapidly find and repair lesions in DNA in the midst of million-fold excesses of undamaged DNA” and could also affect the ability of drugs to recognize specific DNA sites.
Biophysicist and Howard Hughes Medical Institute investigator Barry Honig of Columbia University and coworkers emphasized the importance of shape in the recognition process. “Proteins utilize differences in DNA shape when they recognize specific DNA sequences,” Honig said. He and his coworkers discovered that arginines on protein surfaces recognize “deviations from an ideal B-form double helix, such as variations in the width of the minor groove”—a new mechanism by which proteins recognize duplex DNA.
“We looked at crystal structures of all known complexes of proteins with DNA and analyzed them computationally,” he said. The analysis showed that “the sequence of DNA affects the width of the minor groove of DNA, that width affects electrostatic potential, and that variations in potential are recognized by arginine.”
For many years, people thought of DNA as just a linear sequence of letters that proteins recognized when they bound to specific sites, Honig explained. “Now we’re restoring DNA to the status of a molecule. Instead of just a linear sequence, it actually has nuanced shape. That shape varies with sequence and is used in binding specificity.”
Biophysicist and informatics specialist Tom Tullius of Boston University and coworkers also study local sequence-dependent effects on the structure of DNA and how that influences recognition. They have made extensive use of hydroxyl radical cleavage, a technique in which DNA is cleaved more or less readily depending on the degree of exposure of DNA residues at specific sites. “This chemistry gives you an indirect image of the shape of the grooves of DNA,” Tullius said. He and his team extrapolated that shape information computationally to the entire human genome and published the information in an online database called Orchid.
The researchers found that they could predict differences in the affinity of neomycin for different major-groove sequences on the basis of differences in the calculated structure of DNA at those sequences. In a study on major-groove binding of a protein called Zif268, they likewise found a strong connection between binding affinity and sequence-dependent DNA shape, even though protein binding is harder to analyze than small-molecule binding. And in a collaboration with Honig’s team, Tullius and coworkers found that electrostatic potentials in the minor groove of DNA can also be predicted with hydroxyl radical cleavage patterns.
The ability to calculate minor and major groove structure “allows us to map electrostatic potential throughout the genome computationally, without the need for high-resolution X-ray structures” that would otherwise be required, Tullius said. “We are quite interested in the idea of making these maps and using them to understand places in the genome that would be favorable for certain kinds of proteins to bind to.”
The maps “could also help explain why nucleosomes are positioned at specific spots on genomic DNA,” he said. The mechanism behind nucleosome positioning “is an important problem of widespread interest these days, and there is a lot of controversy and discussion about it,” he added. “We think shape and other physical properties of DNA are going to be important” in understanding nucleosome placement.
Tullius said he hopes his work will also make it easier “to design small molecules that recognize shapes of DNA with more specificity than just sequence” as potential drugs. “It’s a way to get beyond writing down a string of letters and thinking that that represents DNA. We need to start to think about nuances of DNA shape as being really important to understanding protein and small-molecule binding to DNA.”
Advertisement
Arya pointed out that in the past a lot of people in the field of DNA recognition “have focused on targeting sequences, but what’s becoming clearer now is that structure matters a lot, too. Different sequences can potentially have the same structure. An incoming small molecule or protein likely does not care what the sequence is, because DNA shape is an underlying factor in the attraction of DNA to an incoming ligand. Approaches that allow people to look at the shape of DNA are catching on finally. That’s one direction I think the field is going and should go.”
Nevertheless, he added, “the real goal should be to keep shape and sequence together. Only then will you get specific small-molecule binders.”





Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter