Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Activation Of Protein Tags
Enzymology: To prepare biological labels for attachment, E1 enzymes dramatically remodel themselves
by Stu Borman
February 22, 2010
| A version of this story appeared in
Volume 88, Issue 8

In a tour de force chemical, structural, and mechanistic study that took five years, researchers have solved a long-standing mystery in a Nobel Prize-winning field of research—they have shown how E1 enzymes activate ubiquitin and related proteins to tag other proteins.
The study could have implications for drug discovery because ubiquitin and ubiquitin-like proteins have regulatory roles in development, immunity, cancer, and other processes, and E1s help catalyze their attachment to other proteins so they can carry out these roles.
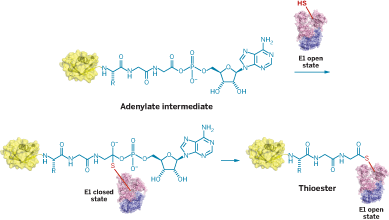
How E1 enzymes work is well-known in many respects, but researchers hadn’t been able to decipher how a ubiquitin or ubiquitin-like substrate is transferred between an E1 enzyme’s two active sites. It is known that an adenylated substrate, bound to an E1 enzyme’s adenylation site, forms a thioester link with a key cysteine at its other site, the Cys domain. But the sites are about 30 Å apart—about 10 times the distance needed for such a transfer to occur.
Derek S. Tan and Christopher D. Lima of Sloan-Kettering Institute, in New York City, and coworkers have now solved that problem by designing analogs of the ubiquitin-like protein SUMO that freeze the E1 enzyme during adenylation and thioesterification. They report their findings in two recent papers (J. Am. Chem. Soc. 2010, 132, 1748; Nature 2010, 463, 906).
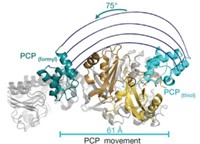
Structures obtained at the two frozen points, plus biochemical and mutational evidence, reveal that SUMO is adenylated by E1 while in an “open” conformation. The enzyme then rearranges to a “closed” conformation to catalyze thioesterification. In the process, the Cys domain turns nearly halfway around, the catalytic cysteine moves 34 Å closer to SUMO, six of E1’s α-helices become disordered or melt into nonhelical “loops,” and half of the residues in the adenylation active site are replaced. The mechanism is believed to apply to other E1s, too.
This “landmark” work “resolves how the transfer is made between the two active sites, which was not obvious from previous structures,” says Arthur L. Haas of Louisiana State University Health Sciences Center School of Medicine, in New Orleans. With Nobel Laureates Avram Hershko and Irwin A. Rose, Haas discovered E1’s basic mechanism and two active sites in 1982. “The clever development of analogs is amazing,” he says. “This is one of the most extreme examples of a massive protein rearrangement, highlighting the plasticity of some proteins.”
“It is extraordinary work, not only for what it provides the ubiquitin field, but also because it helps establish an exciting new paradigm of protein structure,” says Brenda A. Schulman, an expert on ubiquitin-like proteins at St. Jude Children’s Research Hospital, in Memphis. “The old view was one protein domain and one structure. The new work shows a protein adopting radically different structures to mediate distinct functions.”
An E1-targeted agent, MLN4924, is currently in human clinical trials. Millennium Pharmaceuticals Director of Biochemistry Larry Dick says Tan and Lima’s work reveals E1’s “intricate dance” and “provides a structural framework to understand the novel mechanism of MLN4924, which we just published” (Mol. Cell 2010, 37, 102).
Lima, Tan, and coworkers believe the mechanism could aid the search for new drugs. E1 conformations “along the path through adenylation and thioesterification could be targeted for therapeutic intervention,” they write in Nature.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter