Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Synthetic Strollers
Inspired by nature, scientists build walking molecules that stay on track
by Sarah Everts
March 21, 2011
| A version of this story appeared in
Volume 89, Issue 12

Walking with two feet is part of what distinguishes humans from other mammals, so perhaps it’s not surprising that scientists are keen to build two-legged molecules that stroll around with a similar gait.
Nature beat scientists by a few billions years with the evolution of the walking protein motors called myosin, kinesin, and dynein. These biological walkers have two feet that are propelled forward by cycles of conformational changes in multiple protein domains, all powered by adenosine triphosphate. Essential roles for these walking motors abound: They pull chromatin apart during cell division, move organelles such as the nucleus around the cell, and drag cargos of neurotransmitters up and down the axons of neurons.
Over the past decade, researchers have tried playing catch-up with nature. Instead of using proteins as nature does, scientists built the first synthetic walking motors out of complex three-dimensional assemblies of DNA. More recently, small-molecule-based motors that are 1,000 times smaller than their DNA counterparts have also taken their first steps. Scientists are even starting to prod a few DNA-based synthetic walkers to do some work—such as moving cargo.
Although synthetic walkers are still in the early stages of development, researchers hope that one day they will be as valuable to the manufacture of drugs and materials as biological walkers are to the healthy function of human cells.
When chemists and physicists started to think about building synthetic walkers in the early 2000s, they couldn’t work from the molecular blueprint of biological walking motors because the complex mechanisms by which nature propels protein motors forward was not understood in atomic detail.
Instead, biological walking motors delivered conceptual inspiration to those wishing to build synthetic versions, in particular regarding how to saunter at the molecular scale, where random Brownian motion plays a much more significant role than the inertia and momentum that help propel humans forward.
A case in point: In the macroscopic world, “my telephone is just going to sit on the table unless I give it some kinetic energy and knock it off,” says David A. Leigh, who develops small-molecule walkers at Scotland’s University of Edinburgh. “Whereas at the molecular level, everything is constantly moving due to Brownian motion. The walking motor doesn’t need to start moving because it’s already moving. What you need to do at the molecular level is to control the directionality of that motion and get rid of the motions you don’t want.”
Brownian motion is so strong that both biological and synthetic walking motors can’t lift both feet at once—one foot must always be tethered to a track—otherwise, the walker will float off the track and disappear into the solution. Molecular walkers, unlike their human tightrope counterparts, cannot use gravity to remain attached to their track, Leigh says.
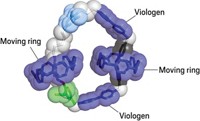
Instead, the first molecular walkers made of DNA used base pairing to stay tethered to their DNA tracks. These walking motors are composed of multiple DNA strands that self-assemble into a walking motor by means of DNA hybridization, the same way double-stranded DNA in our nucleus stays partnered up. Each leg of a DNA walker has a dangling single strand of bases—the so-called foot—that hybridizes with single-stranded footholds that point upward out from DNA double-helix tracks.
As a DNA walker steps along such a DNA track, the foot strands sequentially “hybridize and unhybridize” to move forward, explains William Sherman, a physicist at Brookhaven National Laboratory. Sherman designed the first such walker in 2004 when he was a postdoc with New York University chemist Nadrian C. Seeman.
Sherman and Seeman’s walker moved along DNA-based tracks in inchworm fashion, the leading leg always staying forward, and back leg catching up but never stepping in front of the leading leg (Nano Lett., DOI: 10.1021/nl049527q). That same year, a team led by Niles A. Pierce at California Institute of Technology reported a similar walker that used many of the same concepts but could step like humans, swinging the back leg ahead of the front leg (J. Am. Chem. Soc., DOI: 10.1021/ja047543j).

These initial strategies relied on a so-called DNA fuel strand to power the walkers’ steps. A DNA fuel strand has a sequence of bases that interfere with the base pairing between the track’s footholds and one of the walker’s feet, because the fuel strand hybridizes better with either the foothold or the foot than with the foothold and the foot together. This interference releases the foot from its foothold. The now-released foot can swing forward to seek out the next foothold, thereby taking a step.
Dozens of DNA walkers now exist. Some have more than two legs; others use alternatives to fuel strands to stimulate a step forward. For example, some researchers use protein enzymes that can cleave DNA to cut connections between trailing legs and the track so that the foot can swing forward. Other researchers have devised hybrid DNA and RNA walkers that can themselves chemically cleave connections between their own trailing legs and the track.
More recently, scientists have started to fashion walkers out of small molecules. In late 2009, the first such walker—based on a small, 21-atom molecule—was reported by Max von Delius, Edzard M. Geertsema, and Leigh (Nat. Chem., DOI: 10.1038/nchem.481). The two-legged walker has one sulfur and one hydrazide foot and walks on a track composed of alternating benzaldehyde and benzylic disulfide footholds. Under acidic conditions, the sulfur foot is tethered to the track by means of a disulfide bond, and the hydrazide foot can step forward powered by Brownian motion. When the researchers push the solution to basic conditions, the hydrazide foot forms a covalent bond with the benzaldehyde foothold. By using a redox reaction to sever the disulfide bond, the walker can step preferentially in one direction instead of hopping back and forth in place.
Leigh’s team has since added to the walker a stilbene moiety whose stereochemistry can be toggled by light. This switch in stereochemistry, coupled with changes in pH, determines the walker’s preferred strolling direction.
Next up, researchers developing synthetic walkers are trying to make them walk “further, faster, with more sophistication,” says Andrew J. Turberfield, a physicist at Oxford University. Indeed, synthetic walkers’ stamina still pales in comparison to their biological forerunners. For example, the biological walker kinesin typically takes about 100 steps. The record for DNA walkers is just 16 steps, and that for small-molecule walkers is a mere four steps.
Researchers are also starting to build walkers with some work ethic. Last May, Seeman and his colleagues reported a DNA walker that was able to move cargoes of gold nanoparticles, a first step toward the goal of “coordinating autonomous walkers with the operation of other nanomechanical devices” to make miniature assembly lines, Seeman says (Nature, DOI: 10.1038/nature09026).
Also last year, Harvard University’s David R. Liu reported a DNA-based walker that could perform basic organic synthesis as it stepped forward (Nat. Nanotechnol., DOI: 10.1038/nnano.2010.190). The series of three amine acylations performed by the walker is “an early, and still modest, step toward integrating DNA-based devices with real-world chemistry applications such as synthesis, screening, and sensing,” Liu says. “No one who is simply concerned with making a particular tripeptide now would do so using our walker. But we hope that this study, and others like it, will steadily increase the functionality of these devices so that eventually they will be able to bring truly useful capabilities to the table.”



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter