Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
An Alternative Methylation Route
ACS Meeting News: Details of bacterial enzymes’ unusual methylation step may help fight antibiotic resistance
by Amanda T. Yarnell
April 18, 2011
| A version of this story appeared in
Volume 89, Issue 16

Two bacterial enzymes that install methyl groups on ribosomal RNA do so by a surprising mechanism, according to two independent groups reporting at last month’s American Chemical Society meeting in Anaheim, Calif. Because one of these methylations helps bacteria evade antibiotics, learning more about how these enzymes work may guide those hoping to sidestep antibiotic resistance.
Methylation of RNA typically involves transferring a methyl group from the common methyl donor S-adenosylmethionine to an RNA base. But the enzyme Cfr and its close cousin RlmN instead assemble a methyl group on their ribosomal adenosine substrate using only a methylene fragment from S-adenosylmethionine, reported Squire J. Booker of Pennsylvania State University and Danica Galonić Fujimori of the University of California, San Francisco. The enzymes are thus better described as methyl synthases, rather than the methyltransferases they were expected to be when Fujimori and coworkers first studied them last year (C&EN, March 29, 2010, page 9; J. Am. Chem. Soc., DOI: 10.1021/ja910850y).
Booker, graduate student Tyler L. Grove, and coworkers recently published their work in Science (<a href="http://dx.doi.org/10.1126/science.1200877" target="_blank">DOI: 10.1126/ science.1200877</a>); Fujimori and postdoc Feng Yan’s work recently appeared in Proceedings of the National Academy of Sciences USA (DOI: 10.1073/pnas.1017781108). In Anaheim, Booker and Fujimori described their work in separate sessions in the Division of Biological Chemistry.
Both Booker and Fujimori suggested Cfr and RlmN may have seized upon this unusual pathway because both enzymes are charged with the demanding task of methylating nonnucleophilic centers. Most methyltransferases target nucleophilic centers, according to enzymologist Perry A. Frey of the University of Wisconsin, Madison. The difficulty inherent in methylating adenine’s nonnucleophilic C2 and C8 “requires a new chemical mechanism for methyl transfer by S-adenosylmethionine,” Frey explained.
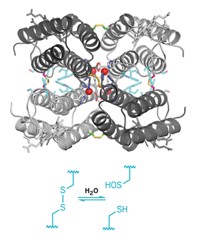
On the basis of isotopic labeling data, the two groups propose different mechanisms for how the enzymes work. A notable difference in their proposals is whether a cysteine in the active site might serve to transfer the methylene fragment from S-adenosylmethionine to the adenine substrate. Adding weight to Booker’s proposal that it might, Amie K. Boal, a postdoc in Amy C. Rosenzweig’s group at Northwestern University, presented an unpublished 2.05-Å crystal structure of RlmN at the meeting. The structure, solved in collaboration with Booker’s group, contains a methylated cysteine near S-adenosylmethionine’s binding site, she reported.
The adenine that both enzymes methylate is at the catalytic heart of the bacterial ribosome, where proteins are built. Although the methyl group that RlmN installs serves to optimize normal ribosomal function, the one that Cfr appends makes bacteria resistant to certain antibiotics. Among the antibiotics rendered inactive by Cfr’s handiwork are the oxazolidinones, a class of synthetic drugs currently used to treat multi-drug-resistant bacteria. Understanding the mechanism by which Cfr confers resistance to these drugs could teach drug designers how to outwit it, Fujimori suggested.
What’s more, Booker said, the mechanism of these enzymes might inform chemists’ continuing efforts to functionalize unactivated carbons.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter