Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Tagging Cholesterol
Chemical Biology: Click chemistry helps label cholesterylated proteins
by Sarah Everts
January 20, 2011
| A version of this story appeared in
Volume 89, Issue 4

Health-conscious folks typically keep a close watch on cholesterol levels, as do researchers studying how this molecule's attachment to proteins can play a role in lung and pancreatic cancer. Now researchers in the U.K. have developed a strategy for monitoring posttranslational cholesterylation in living cells. The new tool will be used to explore cholesterol's known role in certain cancers, when it is attached to a protein called Sonic hedgehog (Chem. Commun., DOI: 10.1039/c0cc04710d).
The technique will also permit scientists to find out how widespread this difficult-to-study posttranslational modification is in cells. Cholesterylation is one of a growing number of posttranslational modifications, such as phosphorylation and acetylation, that control the activation of signaling networks in biological cells.
A team of researchers led by Edward W. Tate at Imperial College London first attached an azide probe to cholesterol's C22–26 chain—as far away as possible from the 3-OH group in cholesterol that attaches to proteins. They fed the modified cholesterol to cells for use in posttranslational modification.
They then built a reporter molecule that attaches to the cholesterol tag via a "click chemistry" reaction, azide-alkyne Huisgen cycloaddition. The reporter molecule also possesses a fluorescent dye that permits imaging and a biotin moiety that enables purification.
The only other option currently available for studying cholesterol posttranslational modifications, radiolabeling, "is hazardous, expensive, time-consuming, not particularly sensitive, and perhaps most important of all, inflexible. A radiolabel just provides a readout on a scintillation counter" instead of the new technique's imaging and purification capabilities, Tate says.
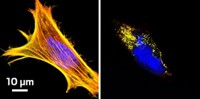
The team went on prove that their technique could tag cholesterylated Sonic hedgehog in cancer cell lines. In doing so, they also discovered that a potpourri of other proteins that were not known to be modified by cholesterol are in fact tagged by the molecule.
The fact that Tate's team "observed so many sites of cholesterol modification in cell proteomes beyond the known Sonic hedgehog protein argues that cholesterylation may be a pervasive means of altering the structure and function of proteins," comments Benjamin Cravatt, a chemical biologist at Scripps Research Institute. It's "a great example of applying bioorthogonal probes to broadly profile an intriguing—and understudied—posttranslational modification."
In addition to finding more examples of cholesterylation in the proteome, the labeling method could be used "to screen drugs that act to inhibit protein cholesterylation directly in living cells, providing a novel approach for cancer therapy," Tate says. "The ability to label protein cholesterylation by fluorescence is unique and may also allow us to track protein cholesterylation over space and time using fluorescence microscopy."


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter