Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Close-Up View Of Protein Folding
Theoretical and experimental studies reveal steps and intermediates in intricate process
by Elizabeth K. Wilson
October 31, 2011
| A version of this story appeared in
Volume 89, Issue 44
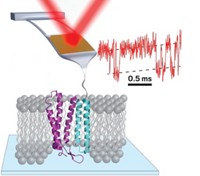
Two new studies—one computational, one experimental—examine the folding of individual proteins, finding that in many cases, repeated foldings tend to move along a prescribed pathway rather than taking many different possible routes. They also find that the folding processes are even more complex than previously thought, with intricate intermediate steps playing important roles (Science, DOI: 10.1126/science.1208351 and 10.1126/science.1207598). Kresten Lindorff-Larsen and colleagues at D. E. Shaw Research used the company’s supercomputer, Anton, to perform folding simulations of 12 proteins ranging from 10 to 80 residues long. The proteins begin to assume their biologically active shapes relatively early in the folding process, they found. Meanwhile, Matthias Rief and colleagues at Germany’s Technical University of Munich attached each end of a single calmodulin protein to tiny glass beads. The beads could then be manipulated with laser beams, allowing the group to probe the protein as it was stretched and unstretched. The identification of defined intermediates “highlights the power of single-molecule techniques,” Tobin R. Sosnick and James R. Hinshaw write in an accompanying perspective.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter