Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Merging Metals Into Proteomics
Tackling the systemic study of metalloproteins
by Jyllian Kemsley
December 12, 2011
| A version of this story appeared in
Volume 89, Issue 50

To understand how proteins work in living organisms, more and more researchers are probing the complex interactions of huge ensembles of proteins as they act together in cells. Many studies involve all the proteins found in cells, tissues, or organisms, collections known as proteomes. The systematic study of those proteins—their structure, function, localization, and modifications, as well as how they change in response to different stimuli—is called proteomics.
An essential subset of the proteome is the metalloproteome, consisting of all the proteins that contain or interact with metals when they do their jobs. Estimates indicate that 9% of eukaryotic proteins bind zinc, 33% of all proteins bind various metals, and 40% of all enzyme-catalyzed reactions involve metals, including Mg, Zn, Fe, Mn, Ca, Co, Cu, Ni, Mo, W, Na, K, and V.
Because the metalloproteome is so important, scientists have devised innovative techniques to better understand its complexities, including not only conventional experimental strategies but also bioinformatics techniques. Using those methods, they have begun to systematically catalog the roles of various metals in protein function.
Bioinformatics is necessary because traditional experimental techniques to purify and investigate individual enzymes are slow and difficult, says Ivano Bertini, a chemistry professor at the University of Florence, in Italy. And just as biologists generally now take advantage of gene and protein databases for insights into how biological systems work, he adds, “we should have something to target metal ions in biology.”
One of the challenges in using information technology to study metalloproteomes, however, is that metal-binding sites in proteins are three-dimensional and don’t always follow a set linear amino acid sequence. Instead, ligands may come from different loops of a protein, different subunits, or even two different proteins, making it difficult to use simple sequence searches to look for protein homology.
In some cases the problem can be addressed by looking for what Bertini calls metal-binding patterns. By mining the Protein Data Bank for structures of known metalloproteins, Bertini, University of Florence chemistry researcher Claudia Andreini, and colleagues determine arrangements of metal ligands along peptide chains and how the ligands bind to metal-binding domains. For a metal bound to three histidine (H) ligands, for example, a metal-binding domain might look something like HXHX60H, where X is amino acids other than histidine.
Bertini, Andreini, and coworkers used that approach to look for zinc-binding proteins in the proteomes of bacteria, archaea, and eukaryotes. From the Protein Data Bank, they found 744 zinc-binding patterns, which they then used to search the proteomes. They found that, on average, 4.9% of bacterial, 6.0% of archaeal, and 8.8% of eukaryotic proteomes are composed of zinc proteins (J. Proteome Res., DOI: 10.1021/pr0603699).
Examining the function of those proteins, the researchers found that most prokaryotic zinc proteins perform some kind of enzymatic catalysis, and two-thirds of them have eukaryotic homologs. Eukaryotic proteomes add to those catalytic enzymes a cohort of zinc proteins that regulate DNA transcription—principally zinc-finger proteins, which likely evolved to meet the complex needs of cell compartmentalization and differentiation in eukaryotes.
More recently, Bertini and Andreini have expanded their database-mining approach to look at 3-D representations of metal-binding sites, focusing specifically on a “minimal functional site” that includes not just a metal and its ligands but everything within a 5-Å radius of the metal ligands. The reactions catalyzed by metalloenzymes are not governed by metal and ligand identity alone, but also by the geometry and environment of the active site, Bertini notes. The additional structure information is therefore critical for predicting function. Two proteins with different overall structures but the same metal functional sites likely do similar chemistry, possibly through convergent evolution of unrelated proteins. In contrast, different metal functional sites found in otherwise similar proteins may have divergently evolved to do different things.
The researchers used minimal functional site analysis to examine nonheme iron proteins, which are proteins that bind iron without using a porphyrin ring. The researchers found that they could group the sites into five structural clusters (J. Mol. Biol., DOI: 10.1016/j.jmb.2009.02.052). A similar effort focusing on zinc sites in proteins yielded 10 clusters of highly similar structures and seven “pseudoclusters” with broadly similar features (PLoS One, DOI: 10.1371/journal.pone.0026325). Comparing and contrasting the active site structures within a cluster can yield clues about how the proteins evolved or how they catalyze their respective reactions, Bertini says. The structural clusters can also help predict the function of uncharacterized proteins.
Bertini and Andreini are also working with Janet M. Thornton of the European Bioinformatics Institute, located in England, to develop a database of the properties and roles of metals in metalloenzyme catalysis. The database is called Metal MACiE; MACiE stands for Mechanism, Annotation & Classification in Enzymes. Metal MACiE pulls together information on the 3-D structure of metalloenzyme active sites as well as a step-by-step description of the reactions they catalyze (Bioinformatics, DOI: 10.1093/bioinformatics/btp256). Although the database has only 188 entries so far, it will be a useful resource for additional systemic studies of metalloenzymes, Bertini says.
But for all that can be learned from mining databases and comparing structural and functional information, the databases are only as good as the information they house, which must originate in experiment. Michael W. W. Adams, a biochemistry professor at the University of Georgia, divides genomes into thirds: We know for certain what proteins are encoded and what the proteins do for about one-third of any particular genome, we have a good idea about another third, and “we have no idea whatsoever” about the remaining third, he says. Of the unknown third, “potentially a lot of them are metal-containing proteins,” Adams says.
Historically, determining the metal content of a protein has often been an experimental afterthought. Scientists purified a protein and then perhaps discovered that it needed a metal to be structurally sound or perform a catalytic function. Some researchers, Adams included, are now reversing that approach by looking for metal content first, by separating proteins on ion-exchange columns or gels and then assaying for metal content. In all cases, researchers note, it is important to purify and characterize proteins in their native forms because proteins that are unfolded or denatured will lose their metal cofactors.
In one set of studies, Adams and colleagues used a combination of inductively coupled plasma mass spectrometry (ICP-MS) to look for metals and high-throughput tandem electrospray ionization mass spectrometry (ESI-MS) to identify proteins in Pyrococcus furiosus, a species of archaea that optimally grows at 100 °C (Nature, DOI: 10.1038/nature09265). After separating P. furiosus biomass through 2-D liquid chromatography—with fractions from one column further separated on a second—Adams and coworkers turned up 343 metal peaks for proteins that variously bound Zn, Fe, Mn, Co, Ni, Mo, W, V, U, and Pb.
Comparing the sequences of the proteins underlying those peaks, they found that 158 of the peaks contained proteins without a previously identified metal-binding domain. Following up on some of those unknowns, Adams and coworkers identified some novel metalloproteins, including one with a new type of Mo site. They also found that the U and Pb peaks most likely came from the organism misincorporating those elements into proteins. Further study of proteins vulnerable to metal misincorporation could reveal mechanisms of metal toxicity, Adams notes.

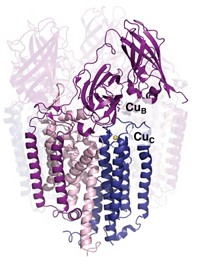
Nigel J. Robinson, a biology professor at Durham University, in England, used a similar approach to determine which proteins predominantly bind Mn2+ or Cu2+ in the periplasm, the space between the cell wall and inner membrane, of the cyanobacterium Synechocystis PCC 6803. Although the microbe requires both metals for photosynthesis, Mn2+ binds weakly and Cu2+ binds strongly to proteins, raising the question of how Mn2+ manages to compete successfully for binding sites.
Analyzing periplasm proteins, Robinson and colleagues turned up something completely unexpected: two proteins with similar structure and metal-binding ligands, one of which contained Mn2+ and the other Cu2+, although both preferred Cu2+ (Nature, DOI: 10.1038/nature07340). Further investigation revealed that the proteins fold and incorporate their metals in different parts of the organism. The Mn-binding protein folds in the cytosol, where researchers believe all copper atoms are already tightly bound to proteins, then gets exported to the periplasm. The Cu-binding protein, in contrast, gets sent to the periplasm unfolded and picks up Cu2+ there. “This highlights the fact that which metals bind to which proteins in vivo is determined by the cell controlling the availability of metals,” Robinson says.
Metalloproteins are not limited to organisms. Viruses, too, may include metals in their proteins. University of Cincinnati chemistry professor Joseph A. Caruso and colleagues have used chromatography and MS techniques to look at bacteriophage λ, a virus that infects and replicates in bacteria. Although the genome and proteome of the virus have been well studied, its metal complement has not. The virus encases its genetic material in a protein cage and then injects it into a bacterial cell to replicate. The protein cage contains a number of cysteine and methionine residues, which are common metal ligands. Caruso and colleagues found that bacteriophage λ incorporates Zn, Fe, Mn, Co, Ni, and Cu, most likely in two proteins in the virus’s cage (Metallomics, DOI: 10.1039/c0mt00104j). Scientists are interested in engineering bacteriophage λ to produce nanoparticles or deliver drugs, and understanding the role that metals play in the virus may further those efforts, Caruso says.
In other application-oriented work, Caruso and colleagues have done metalloproteomic screens for biomarkers to predict disease. In one case, they looked for a particular form of the iron-transport protein transferrin as a signal for leaking cerebrospinal fluid after head injury (Analyst, DOI: 10.1039/c0an00207k). Another study involved identifying possible markers for narrowing of brain arteries after hemorrhagic stroke (Metallomics, DOI: 10.1039/c0mt00005a).
Chromatography combined with MS techniques to study metalloproteins could be improved, says David W. Koppenaal, chief technology officer of the William R. Wiley Environmental Molecular Sciences Laboratory at Pacific Northwest National Laboratory. Right now, combining techniques such as ICP-MS to get metal content and ESI-MS to get protein sequences means doing them separately and correlating the data. Koppenaal’s lab and others are working on technology to use one or more chromatography columns to separate samples, then split the eluent into two streams: one for elemental analysis through ICP-MS and the other for molecular mass analysis through ESI-MS or other techniques.
A simultaneous, integrated approach enables unequivocal and global determinations of metal-protein associations, allowing the researchers to collect “richer and more precise information,” Koppenaal says. One of the projects that Koppenaal and colleagues are working on is investigating the photosynthetic machinery of cyanobacteria. “We’ve done a lot of proteomics work on the system, and now we want to complement that work with better characterization of the protein-associated metals, metabolite-associated metals, and free metals,” Koppenaal says. “Understanding the dynamic among those metal pools is an important part of the whole picture.”
A slightly different approach to metalloproteomics is to separate proteins by 2-D gel electrophoresis and then analyze the gel bands for metals. Norbert Jakubowski, a scientist at BAM Federal Institute for Materials Research & Testing, in Germany, has used gel electrophoresis plus laser ablation ICP-MS to analyze Se enrichment in proteins and to profile cytochrome P450 expression in rat livers (J. Anal. At. Spectrom., DOI: 10.1039/c003889j and 10.1039/c0ja00077a), as well as to look for Cd in spinach.
Advertisement
Plants can take up Cd and other toxic metals from soil, potentially reducing plant growth and contaminating the food supply. Researchers suspected that Cd was displacing other metals in enzymes, rendering them nonfunctional, but they didn’t know which proteins Cd affected. Jakubowski and colleagues analyzed protein extracts of spinach leaves and found that Cd mainly substitutes for Mg in the active site of ribulose-1,5-bisphosphate carboxylase oxygenase, commonly known as RuBisCO, a critical enzyme in carbon fixation and the most abundant protein in leaves (Metallomics, DOI: 10.1039/c1mt00051a).

Other ways to detect metalloproteins in gels include X-ray fluorescence mapping, which allows simultaneous imaging of multiple metals to get identity and quantity. Adding X-ray absorption near-edge structure analysis can also reveal metal oxidation states. Argonne National Laboratory scientist Lydia Finney and coworkers used the X-ray techniques to study the effects of Cr3+ and Cr6+ spiked into blood serum, demonstrating that Cr3+ compounds promoted as nutritional supplements bind to serum proteins and form some Cr6+ species, contrary to marketing claims (ACS Chem. Biol., DOI: 10.1021/cb1000263).
Finney is now collaborating with Worcester Polytechnic Institute biochemistry professor José M. Argüello to investigate metal-trafficking pathways in bacteria by mutating known copper transport proteins. The ability to simultaneously image multiple metals is critically important to the study, she says. “We’re specifically interested in what happens with copper, but we also see big changes in other metals,” Finney says. “Their homeostasis is very intertwined.”
In yet another approach to metal detection, researchers at Delft University of Technology, in the Netherlands, take advantage of an on-site nuclear reactor to enrich protein samples with rare nuclear isotopes, such as 69mZn, 59Fe, 64Cu, 99Mo, and 187W. Biotechnology professor Peter-Leon Hagedoorn and colleagues have dosed Escherichia coli cultures with the isotopes and tracked where the metals go: More than 99% of Cu is located in the protein CueO, a multicopper oxidase involved in Cu homeostasis (J. Biol. Inorg. Chem., DOI: 10.1007/s00775-009-0477-9), and 90% of Fe resides in superoxide dismutase, ferritin, and bacterioferritin (Metallomics, DOI: 10.1039/c1mt00154j). Under Zn stress, E. coli shuttles Zn to ZraP, which Hagedoorn proposes is a novel prokaryotic Zn storage protein that scavenges Zn in the periplasm. His group has also looked at how P. furiosus distinguishes between chemically similar Mo and W in its metalloproteome (J. Bacteriol., DOI: 10.1128/JB.00270-10).
One additional method to study metalloproteomes originated with the U.S. Protein Structure Initiative (PSI) to determine the 3-D structures of all proteins. The initiative organizes sequences into families and then solves the structures of selected representatives of each family. At Brookhaven National Laboratory, Case Western Reserve University proteomics professor Wuxian Shi and colleagues developed a high-throughput X-ray absorption spectroscopy (HTXAS) technique to identify and quantify metals in nearly 4,000 proteins selected for PSI analysis.
The combination of crystal structure, HTXAS information, sequence homology comparisons, and bioinformatics mining of protein sequences for likely metal ligands helped clarify the metal-binding sites in the proteins (Genome Res., DOI: 10.1101/gr.115097.110). Although the project previously screened all proteins by HTXAS, in the next phase the scientists will analyze only those proteins that have been crystallized.
For all the work that has gone into studying metalloproteins, there is still much to learn, Adams says, pointing again to the one-third of the genome about which “we don’t have much of a clue.” It took roughly 100 years to get the knowledge that we have now, he notes. How long it will take to close the gap, even with new, high-throughput proteomics, remains to be seen.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter