Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
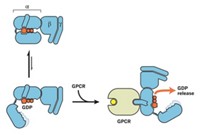
Ion Channel In Motion
Computational Biochemistry: Simulation tour de force visualizes protein’s opening and closing
by Stu Borman
April 16, 2012
| A version of this story appeared in
Volume 90, Issue 16

One of the most extensive biomolecular simulations ever has allowed researchers to visualize the opening and closing of a voltage-gated potassium ion channel for the first time.
Voltage-gated ion channels in cell membranes help propagate nerve impulses, time heartbeats, and synchronize muscle contractions. The findings could thus aid drug design for heart disease, paralysis, migraine, and other conditions caused by ion-channel malfunctions. The work also shows that new levels of computer power are becoming available to study biomolecules.
Structures of voltage-gated ion-channel open states have been obtained, and many other studies have revealed much about channel behavior. But closed-state structures have remained elusive, making it difficult to nail down the channels’ overall mechanism.
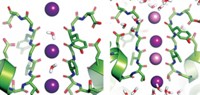
Now, computational biochemist David E. Shaw of D. E. Shaw Research, in New York City, and Columbia University and coworkers including Morten Ø. Jensen have used a customized computer called Anton to perform all-atom calculations on a long-enough timescale to simulate ion-channel opening and closing (Science, DOI: 10.1126/science.1216533). The study was funded by Shaw Research and not supported by government grants.
The study of a system of more than 100,000 atoms was made possible by Anton’s ability to perform molecular dynamics simulations about 100 times faster than those carried out by any other computer. The longest simulation time in the new study is 230 microseconds, whereas comparable simulation times on other computers have been about 10 microseconds at most.
To make their computationally demanding simulations of channel opening and closing fast enough to be practical, the Shaw group applied membrane voltages several times higher than normal. That maneuver could spark controversy about whether the simulations elicited realistic channel behavior.
S4 helices on each of the channel’s four voltage-sensing domains are the main moving parts. The simulation shows them twisting as they open and close the channel. The group also simulated the activity of a channel with a known heritable mutation and proposed a mechanism for its aberrant ion flow, which is believed to cause heartbeat irregularities and neurological problems.
“Amazing!” said ion-channel expert Frederick J. Sigworth of Yale School of Medicine after viewing a movie of the normal process. “It’s like seeing for the first time something that until now has existed only in imagination. There are going to be things shaken out about whether Shaw and company got the details right, but it’s very impressive that they were able to put together a pretty convincing physical system and let it run.”
The new findings agree with a general consensus about the mechanism that has developed in the past couple of years, Sigworth and others tell C&EN. However, researchers disagree or are uncertain about some mechanistic details, such as how much S4 moves and whether or not it twists. Shaw’s simulation could help resolve such points of contention.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter