Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Synthesis
Precatalytic Intron Structure Solved
Pre- and postcatalytic structures of a self-splicing RNA molecule (the group II intron from a bacterium) are similar
by Celia Henry Arnaud
April 16, 2012
| A version of this story appeared in
Volume 90, Issue 16
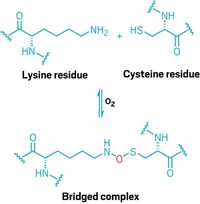
The first crystal structure of the precatalytic state of a group II intron has been solved (Nat. Struct. Mol. Biol., DOI: 10.1038/nsmb.2270). Previous structures of these self-splicing RNA molecules, which are thought to be evolutionary predecessors of the spliceosome, have captured them either during or after the splicing process, but never before splicing starts. Navtej Toor of the University of California, San Diego, and coworkers obtained a 3.65-Å crystal structure of the group II intron from the ocean sediment bacterium Oceanobacillus iheyensis in its precatalytic state. To trap this state, the researchers deactivated the catalytic site with a single-point mutation. The structure shows that the 5ʹ splice site has a kinked structure before catalysis, which had been predicted. The researchers propose that this structural distortion of splice sites is a general mechanism inherent to all RNA-splicing reactions. They also modeled the probable position of the 3ʹ splice site and used this information to propose a mechanism for the entire splicing pathway. Their model suggests that the active site can accommodate both splicing steps with relatively small conformational alteration.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter