Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Materials
More Than Just A Smiley Face
Researchers struggle to find practical applications for nanoscale patterning technique
by Bethany Halford
July 9, 2012
| A version of this story appeared in
Volume 90, Issue 28

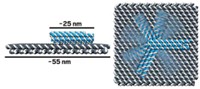
If people recall only one thing from Paul W. K. Rothemund’s 2006 paper introducing DNA origami to the world, it’s the smiley faces. Looking at the 20-odd 100-nm visages grinning in one of the report’s atomic force microscopy (AFM) images, one can’t help but wonder if there was ever a more cheerful chemical picture.
“Everybody remembers the smiley faces,” says Nadrian C. (Ned) Seeman, the New York University chemistry professor considered to be the founding father of DNA nanotechnology. Decades earlier, Seeman spent years figuring out how to forge cubes and truncated octahedrons out of small lengths of DNA. Later, Rothemund, a computer scientist at California Institute of Technology, developed a clever design to create shapes out of DNA that were 10 times as complex as anything anyone had ever made from the biopolymer (Nature, DOI: 10.1038/nature04586).
What Seeman found most impressive in Rothemund’s work was the sheer scale of it. “It was really quite extraordinary,” he says.
In DNA origami, one long single strand of DNA from the M13 bacteriophage—roughly 7,000 nucleotides long—gets folded into a predetermined shape with short, so-called staple strands of DNA. Since Rothemund’s original report, researchers have gone on to use DNA origami to create DNA dolphins with movable tails, three-dimensional polyhedrons, and boxes with locks, among other things.
But back in 2006, Lloyd M. Smith, a chemistry professor at the University of Wisconsin, Madison, remarked in a commentary on DNA origami that “the barrier we have to surmount next is to deploy our knowledge to develop structures and devices that are really useful.”
Six years later, DNA origami experts are still grappling with that barrier. “I think the main challenge facing DNA origami is demonstrating useful applications of the technology,” says Shawn M. Douglas, a technology development fellow who works in DNA nanotechnology at Harvard University’s Wyss Institute for Biologically Inspired Engineering.
“If you’re interested in making objects out of DNA, there’s an incredible amount of science there,” Rothemund tells C&EN. There’s plenty to glean from digging into the basic thermodynamics, biophysics, kinetics, and folding of DNA origami, he says, but the technique’s ability to create nanoscale patterns is where the action is. “We can pattern easily at the 50-Å scale; now what do you do with that?” Rothemund wonders.
“In my mind, the application that’s really going to take off in the next 10 years is using DNA to study other biological objects,” Rothemund continues. For example, he says, one could use DNA origami to create a jig that holds proteins or other molecules in a certain arrangement so they can be studied.
“There are lots of questions we have in basic science about how molecules interact with each other, what forces are involved, or how cells interact with each other and how things are arranged on the nanoscale,” Douglas says. “DNA nanotechnology has great potential as a tool to build devices to start answering questions about how biomaterials are arranged in 3-D or how they interact with one another in ways that were previously inaccessible.”

Hendrik Dietz, a DNA nanotechnologist at the Technical University of Munich (TUM), in Germany, is working on building such tools. For example, he envisions using the origami method to make DNA tweezers that could grab on to a single protein molecule. Such a device could pull apart protein molecules and reveal their inner workings, he says.
Along with TUM colleague Ulrich Rant and coworkers, Dietz recently used DNA origami to create customized gatekeepers for solid-state nanopores (Angew. Chem. Int. Ed., DOI: 10.1002/anie.201200688). The DNA structures have small apertures at their centers. Those structures sit on the nanopores and control the passage of macromolecules.
Because Dietz and Rant’s team can control the DNA sequence at the apertures, they can create structures that latch on to specific sequences of DNA and guide them through the pores. This, Dietz says, suggests they “could rig the device to detect all kinds of molecules in complex solutions,” such as disease markers.
Another example of how DNA origami is being used to study biomolecules comes from researchers at Kyoto University, in Japan. A team led by Masayuki Endo and Hiroshi Sugiyama has used DNA origami to create a device that lets them observe the real-time behavior of single DNA-binding enzymes by high-speed AFM at molecular resolution. This gives them a direct glimpse at how the enzyme is behaving, Endo and Sugiyama tell C&EN, as opposed to the indirect-fluorescence-microscope-based single-molecule analyses traditionally used.
Endo and Sugiyama’s setup features a picture frame made from DNA origami with two lengths of DNA strung across it. One strand is taut and the other is loose enough to be bent by the studied enzymes.
Using the device, dubbed a DNA nanochip, they note that DNA methylation enzymes and DNA repair enzymes work better with the looser strand. This, Endo and Sugiyama say, indicates the importance of structural flexibility in the DNA for association with these enzymes (J. Am. Chem. Soc., DOI: 10.1021/ja907649w; Angew. Chem. Int. Ed., DOI: 10.1002/anie.201003604).
As exciting as these applications may be to researchers who study single biomolecules, those working in DNA origami concede that they’re unlikely to impress people outside the scientific community. One area that Harvard’s Douglas thinks DNA origami, as well as other biomolecule construction techniques, will be more attention grabbing is in creating devices that are as sophisticated as viruses.
To that end, he, along with Wyss colleagues George M. Church and Ido Bachelet, used DNA origami to build a nanorobot capable of delivering therapeutic cargo specifically to cancer cells (Science, DOI: 10.1126/science.1214081; C&EN, Feb. 20, page 8). Creating the robot was a proof-of-concept experiment, Douglas says. To actually work as a drug delivery device, the structure would need to be optimized and likely redesigned.
Aside from finding the perfect application, the field of DNA origami faces technical hurdles. “Making shapes is one thing,” TUM’s Dietz says, “but making a functional shape is yet another challenge.” To make functional DNA molecules, he notes, one has to be able to make the DNA structures in high yield and with very high quality. So far, he says, that’s been tough.
Another problem, Rothemund points out, is putting the DNA origami structures, which are made in solution, onto a surface. “It’s like taking a deck of cards and throwing them on the floor. They go everywhere,” he says. “If you’re trying to wire them into a circuit or study some protein, you have to go find where the DNA origami is on the surface.”
Working with scientists at IBM, Rothemund has made patterned, positively charged surfaces to which the negatively charged DNA origami will stick, but he says the work has been difficult to reproduce. Ideally, he’d like to be able to make a kit that he could get into the hands of researchers who are interested in working with DNA nanostructures. “We want to demonstrate that if you have the power to put DNA origami where you want, you can really build something where you can do some science that you wouldn’t otherwise be able to do,” he says.
Despite the challenges, the scientists who spoke with C&EN think that DNA origami research will find its groove. After all, they say, DNA is an ideal molecule to work with.
“It’s very durable,” Dietz says. “You can take a DNA molecule, put it in water, and let it sit on your desk for a year at room temperature.” Its Watson-Crick base pairing is also extremely reliable, he adds. “If you want a DNA double helix to form, all you have to do is write down complementary sequences and mix the molecules together, and they will form a DNA double helix. You can count on it. There are very few systems where you have this kind of certainty.”
“It’s also chemically versatile,” Douglas points out, “so you can modify it very easily, but it’s also very stable. There’s a reason nature uses it as an information storage molecule in our cells. As soon as we get commercially viable applications, DNA origami will draw a lot of resources and excitement to the field. I’m definitely optimistic about where it will go in the future.”
Rothemund says, “Just wait five years and see what we really can do.”



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter