Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Materials
Halogen Bonding Defined
IUPAC effort sets parameters for structural interaction seen in materials science and biology
by Jyllian Kemsley
August 27, 2012
| A version of this story appeared in
Volume 90, Issue 35

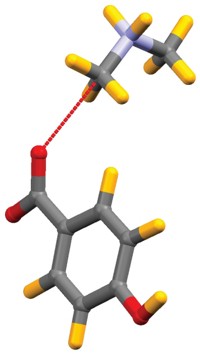
Halogen bonding, akin to hydrogen bonding, is a noncovalent interaction between a positive region on a halogen atom and a negative site, such as a lone pair on a nitrogen or oxygen. Following on the heels of a new definition for hydrogen bonding, an International Union of Pure & Applied Chemistry (IUPAC) committee has now proposed a definition for halogen bonding. The definition will promote recognition of halogen bonding as a key structural element in materials science and biology, scientists working in the field say.
The proposed definition says that a halogen bond is a net attractive interaction between an electrophilic region of a halogen atom on a molecule or molecular fragment and a nucleophilic region of another molecule or molecular fragment. The electrophilic region arises on the halogen because when a halogen makes a covalent bond in a molecule, the electron distribution around the atom shifts a bit toward the bond. That leaves an area of diminished electron density opposite the bond—an area christened as the σ-hole.
The σ-hole results in a region of positive electrostatic potential surrounded by an equatorial band of negative electrostatic potential. The geometry of the potentials around the halogen atom means that, in contrast to hydrogen bonds, halogen bonds are directional, occurring only in line with the halogen’s molecular bond. Even fluorine can form halogen bonds, if it is covalently bonded to sufficiently electron-withdrawing groups, such as in FCOF.
Like the hydrogen bond definition (C&EN, Nov. 22, 2010, page 32), the halogen bond definition lists frequent but not necessarily required characteristics of halogen bonding: that the length of a halogen bond, for example, is less than the sum of the van der Waals radii of the atoms involved, and that formation of a halogen bond may change absorption or nuclear magnetic resonance spectra. “The greater the number of features satisfied, the more reliable is the characterization as a halogen bond,” the definition says.
The definition does not attempt to address the nature of the halogen-bonding interaction. “You can explain all of the character with electrostatics,” says Tim Clark, a chemistry professor at Germany’s University of Erlangen-Nuremberg, although other researchers incorporate additional types of interactions or assert that halogen bonds have some covalent character.
The reason for developing the definition is growing recognition of the roles halogen bonds play in the structure of materials, from inorganic to biological, say IUPAC committee chairs Pierangelo Metrangolo and Giuseppe Resnati, chemistry professors at the Politecnico di Milano, in Italy. Metrangolo and Resnati have explored halogen-bonding effects in liquid crystals, ion recognition and binding in supramolecular chemistry, and light-induced surface patterning of supramolecular polymers.
The area in which halogen bonding is poised to make big changes is in biochemistry. Current modeling software treats halogens as having all-negative electrostatic potential, eliminating the possibility of predicting halogen bonds. Yet many natural protein inhibitors involve halogens, notes P. Shing Ho, a biochemistry professor at Colorado State University. His group and others are working to better define the effects of halogen bonding in biology and to update potential energy functions for modeling.
Because halogen bonds have a strong geometric requirement, “it’s much more difficult in the protein to establish a strong halogen bond than a strong hydrogen bond,” says François Diederich, a professor of chemistry at the Swiss Federal Institute of Technology, Zurich. “But if you find the right halogen bond and establish it, you have made a big gain in affinity and also selectivity.”
Researchers are also looking to explore halogen-like bonds made by elements in groups 16, 15, and 14 of the periodic table. “Covalently bonded atoms in these groups can have two, three, and four σ-holes, respectively, or more if hypervalent,” says Peter A. Politzer, an emeritus chemistry professor at the University of New Orleans.
IUPAC is accepting comments on the halogen bond proposal through the end of November.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter