Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Proteins Made To Order
Design Rules: Principles integrate structure and function from the ground up
by Celia Henry Arnaud
November 19, 2012
| A version of this story appeared in
Volume 90, Issue 46

Efforts to design novel protein catalysts have typically involved modifying naturally occurring proteins for a new purpose. But the resulting proteins are often less than ideal. Now, David Baker of the University of Washington, Seattle; Gaetano T. Montelione of Rutgers University, Piscataway; and coworkers have developed guidelines for designing proteins from the ground up with results that are close to ideal (Nature, DOI: 10.1038/nature11600).
“With this new approach, we can start designing fold and function at the same time,” Baker says. Designing structure and function together, he says, can lead to more active catalysts than existing strategies, which involve starting with naturally occurring protein structures and trying to change their function by embedding new catalytic sites in them.
“For an active site, you really need to have all the catalytic groups in exactly the right place. When you reuse a backbone that evolved for something else, the backbone won’t be placed exactly where you need it to be,” he says. “If you’re building the protein from scratch and you have high-precision control over the position of the backbone, you really can get the geometry exact.”
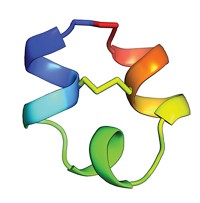
Baker’s team came up with rules that describe the junctions between adjacent structural elements such as α-helices and β-strands. The rules depend on the chirality of ββ units and the orientation of αβ and βα units—characteristics that are dictated by the length of each structural element and the loops that connect them.
Notably, these rules are mostly independent of the amino acid sequences. “A lot can be controlled before you even get to the point of thinking about the details of the sequence, simply by controlling the length of the secondary structure elements and by controlling the length of the turns,” Baker says. Monte Carlo simulations can then identify the best sequence for the chosen backbone.
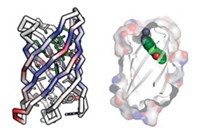
Baker and coworkers designed a variety of proteins using the rules. They then enlisted volunteers using the distributed computing project Rosetta@home to predict the proteins’ lowest energy structures. They chose five proteins whose predicted structure matched the designed structure, expressed them in bacteria, and obtained their structure using NMR spectroscopy. They found that the actual structures also matched the design.
William F. DeGrado, professor of pharmaceutical chemistry at the University of California, San Francisco, calls the rules a “new, comprehensive, and exciting approach for de novo protein design.”
Next up, Baker and colleagues plan to incorporate catalytic sites into the scaffolds they have designed, none of which actually have active sites.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter