Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Driving The Circadian Clock
Researchers home in on the biochemical mechanism of how cells keep time
by Jyllian Kemsley
December 3, 2012
| A version of this story appeared in
Volume 90, Issue 49

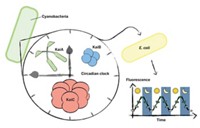
As travelers crossing time zones know all too well, humans and other organisms have circadian clocks, the biological machinery that controls daily cycles of gene expression and physiology. The simplest organism known to possess a circadian clock is a type of photosynthetic bacteria called cyanobacteria, or blue-green algae. The gears of the cyanobacterial clock in Synechococcus and Thermosynechococcuselongatus are three proteins known as KaiA, KaiB, and KaiC, and researchers have recently made several breakthroughs in understanding the biochemical mechanism of exactly how those cyanobacteria keep time.
The recent discoveries are based in part on earlier key work published in 2007 papers. Two research groups determined that ordered phosphorylation and dephosphorylation of two KaiC amino acids are key to cyanobacterial clock oscillation (EMBO J., DOI: 10.1038/sj.emboj.7601832; Science, DOI: 10.1126/science.1148596). One of the groups was led by Takao Kondo, a professor of biological science at Japan’s Nagoya University, and the other by Michael J. Rust, now a professor of molecular genetics and cell biology at the University of Chicago, and Erin K. O’Shea, a professor of molecular and cellular biology at Harvard University.
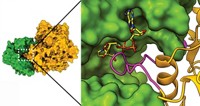
At that point, researchers assumed that each of several phosphorylated and dephosphorylated forms of KaiC had a distinctly different structure that dictated how it interacted with other proteins, says Andy LiWang, a chemistry professor at the University of California, Merced. Scientists thought the clock would be a set of largely static forms, “chunking” from one to another, he says.
Then, in 2009, Vanderbilt University School of Medicine biochemistry professor Martin Egli and colleagues published a set of KaiC crystal structures that incorporated amino acid mutations designed to mimic the different phosphorylation states (PLoS One, DOI: 10.1371/journal.pone.0007529). All of those structures looked essentially the same, raising the question of just what phosphorylation and dephosphorylation accomplished.
But work published last year and this year from LiWang’s lab using nuclear magnetic resonance spectroscopy to study the Kai proteins is painting a new picture of the inner workings of the clock (Proc. Natl. Acad. Sci. USA, DOI: 10.1073/pnas.1104221108 and 10.1073/pnas.1211508109).
In aggregate, here’s the biochemical cycle as it’s now understood: Structurally, KaiC is composed of two homohexamer stacked rings called CI and CII. At the start of the morning, KaiC is unphosphorylated. So-called A-loops of protein on CII exist in dynamic equilibrium between being buried in the CII ring or exposed to its surroundings. KaiA binds to and captures the A-loops in their exposed position, stimulating KaiC over the course of the day to autophosphorylate first a threonine and then a serine residue on CII.
Prior to phosphorylation, the CI and CII rings float a bit apart and the CII ring is flexible. Phosphorylation of the CII serine residue causes the CII ring to become more rigid and stack onto the CI ring. That stacking wedges the CI subunits a bit apart, creating space for KaiB to bind to CI toward the end of the day. Prior to LiWang’s studies, researchers thought that KaiB bound to CII.
As day crosses into night, KaiB binding to CI triggers KaiA to move from CII to CI. Just as KaiA binding to CII promoted phosphorylation during daytime, removing it allows for dephosphorylation overnight. The protein dephosphorylates the threonine first, then the serine. Once the serine loses its phosphate group, CII relaxes and disengages from CI, allowing the CI hexamer to close up and push off KaiB.
Fully dephosphorylated and free of KaiA and KaiB, KaiC is then ready to start the day again.
Even as LiWang’s new mechanism of how the Kai proteins interact with each other upends some previous work, it helps to explain other research. One area is in clock output, or how the circadian clock’s biochemical rhythm gets transmitted to the rest of the cell to regulate events such as gene expression and cell division. A protein known as SasA is key to that output and binds to the CI ring competitively with KaiB. LiWang believes that KaiC ring stacking and unstacking influences SasA interactions so SasA can transmit the circadian phase to other parts of the cell.
A separate area of active research involves how the cyanobacterial clock is regulated and, in particular, reset in response to changes in light. People had thought clock resetting was a complex, regulated pathway, LiWang says, but it turns out to be simpler.
In one study, Rust and O’Shea, together with University of California, San Diego, biological sciences professor Susan S. Golden, demonstrated in 2011 that light-induced resetting of the cyanobacterial clock is associated with changes in cellular amounts of adenosine triphosphate (ATP) and its precursor adenosine diphosphate (ADP) (Science, DOI: 10.1126/science.1197243). If dusk happens early or there is some other light interruption, cyanobacteria stop making ATP, ADP builds up, and the clock moves into nighttime mode. LiWang’s work suggests that the mechanism involves ADP wedging apart the CI ring to promote KaiB binding.
More recently, Golden’s lab has also demonstrated another link between light and clock regulation. Onset of darkness causes oxidation of cellular plastoquinones, which play a key role in photosynthesis. Oxidized plastoquinones bind to KaiA, inhibiting its interaction with CII and promoting dephosphorylation (Proc. Natl. Acad. Sci. USA, DOI: 10.1073/pnas.1216401109). “There is a lot of evidence now that the KaiABC system is taking in information about the metabolic state of the cell to set the clock time,” Rust says.
One issue that remains a mystery is just how the proteins stay tuned to a 24-hour cycle. An extraordinary feature of the Kai protein system is that if you just put the three proteins in a test tube with some ATP, on their own they will set up a 24-hour oscillation.
And the system is remarkably sluggish, at least compared with other protein machinery. “If you think about the time it usually takes for protein-protein collisions or catalysis, this is orders of magnitude slower,” Rust says. “Evolution was trying to make the rate constant be a particular value rather than trying to just make it be fast,” as we would normally expect for enzyme catalysis, Rust says.
Some of that built-in timing hinges on slow CII phosphorylation, although why it is so slow remains to be determined. Another component of timing is likely KaiB, which exists as a tetramer in solution but binds to CI as a dimer and may also have a monomer state. The equilibrium among KaiB tetramer, dimer, and monomer is probably a key component of clock timing, LiWang says.
KaiC is also an ATPase that converts ATP to ADP. That ATPase activity is necessary for clock function, and it is slow. Rust is working to understand the role that it plays in determining clock timekeeping.
The details of the cyanobacterial clock may lead to new insights into the mammalian clock, Egli says. Some researchers believe the human clock runs on some sort of transcription or translation feedback loop, as scientists formerly thought about cyanobacteria. Do other organisms also have biochemical clocks programmed to keep time intrinsically? That’s a study for another day.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter