Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Protein Takes Aim At Celiac Disease
Team redesigns enzyme to stop harmful immune reactions caused by gluten-based peptides
by Stu Borman
December 10, 2012
| A version of this story appeared in
Volume 90, Issue 50

People with celiac disease, who cannot tolerate gluten in their diet, can thank a team of undergraduates for a new entry in the pipeline of oral drug candidates to treat their condition. The students, guided by academic advisers, used computational protein design to considerably boost a peptidase’s ability to attack and disable problematic components of gluten.
The redesigned enzyme resists breakdown by gut enzymes, so it could be an oral medication for celiac disease. The research team has formed a company to test and develop the drug candidate commercially. But the enzyme has many bridges to cross to be shown safe and effective, and it will have to compete with other celiac disease drug concepts, some of which are much further along in the development pipeline.
Celiac disease is an inflammatory reaction to proline- and glutamine-rich peptides in gluten, a glycoprotein in wheat, rye, and barley. Such peptides are hard to digest and cause immune reactions in susceptible individuals, resulting in intestinal damage and symptoms such as cramps, gas, bloating, diarrhea, constipation, anemia, and weight loss. The new enzyme is designed to destroy such peptides in the body after a meal containing gluten before they can do harm.
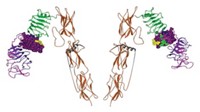
This work was carried out by University of Washington undergrads, advised by UW biochemistry senior research fellow Ingrid Swanson Pultz and assistant professor of biochemistry and chemistry Justin B. Siegel of the University of California, Davis, as part of the International Genetically Engineered Machine synthetic biology competition (J. Am. Chem. Soc., DOI: 10.1021/ja3094795).
To build the drug candidate, the team first identified a natural enzyme that might work well in the acidic environment of the stomach—kumamolisin-As from Alicyclobacillus sendaiensis, a bacterium that grows in acidic soils in Japan. They then used the computational enzyme design tools Foldit and the Rosetta molecular modeling suite, both developed by UW protein design expert David Baker and coworkers, to engineer the enzyme to target immunogenic gluten-based peptides. KumaMax, the group’s name for the engineered enzyme, is 877 times more selective than the native enzyme for peptides containing a proline-glutamine dipeptide motif and 116 times more effective at cutting up one of the major problematic celiac disease peptides.
“We think this enzyme has great potential” for treating celiac disease, Pultz says. In October, she, Siegel, and Baker founded Seattle-based biotech company Proteus Biologics to develop it.
But the designed enzyme has high barriers to overcome. “The gluten-free diet is by far the most effective treatment for celiac disease,” says Alessio Fasano, director of the University of Maryland Center for Celiac Research. “It’s very efficacious and pretty much devoid of side effects. So the bar for gluten-free-diet alternatives is high.”
And nearly 50 clinical trials of other potential treatments have been or are being carried out, he says. The concepts on which they’re based include not only breaking down gluten fragments but also reducing gut permeability to gluten peptides, inhibiting an endogenous enzyme that makes the peptides immunogenic, blocking immune reactions to the peptides, and vaccinating to induce tolerance to them.
“The beauty of the new study” is that the engineered enzyme has an enhanced ability to digest gluten-derived peptides “to an extent I’ve never seen before,” Fasano says. “This holds strong promise in moving the field forward—to help people eat regular food without being harmed by gluten.” But he’s not sure the enzyme would work fast enough in vivo for a celiac disease patient “to eat a Big Mac and not be harmed.”
Brent L. Iverson, a protein design specialist at the University of Texas, Austin, says, “What they’re trying to accomplish is a tall order—put an enzyme in your stomach and have it go after peptides at exactly the right time, before they are absorbed and cause immune problems.”
“Now they have to show that the enzyme is really capable of doing the job under the required circumstances,” says Frits Koning of Leiden University Medical Center, in the Netherlands, whose interests include gluten-degrading enzymes.
“Time will tell if this is a baby step or if they slayed the dragon,” Iverson adds.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter