Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Solving Powder Structures
Spectroscopy: Computational NMR approach deciphers what other methods cannot
by Jyllian Kemsley
November 11, 2013
| A version of this story appeared in
Volume 91, Issue 45
![Crystal structure of 4-[4-(2-adamantylcarbamoyl)-5-ter-butyl-pyrazol-1-yl] benzoic acid, aka AZD8329, a type 2 diabetes drug candidate. Crystal structure of 4-[4-(2-adamantylcarbamoyl)-5-ter-butyl-pyrazol-1-yl] benzoic acid, aka AZD8329, a type 2 diabetes drug candidate.](https://s7d1.scene7.com/is/image/CENODS/09145-notw3-crystalstructurecxd?$responsive$&wid=300&qlt=90,0&resMode=sharp2)
Molecules can crystallize in different forms, which may have variable stability or solubility properties. For materials and pharmaceutical applications that use microcrystalline powders, manufacturers must be able to reliably produce specific crystal forms and have analytical methods available to ensure that they’re doing so.
But such testing is easier said than done. “Solving structures of molecular solids from powder diffraction data is highly challenging, and complementary techniques are desperately needed,” comments Paul Hodgkinson, a chemist at England’s Durham University.
By combining computational work and solid-state nuclear magnetic resonance spectroscopy (NMR), an international team of researchers has pioneered a technique to solve the crystal structure of a small molecule (J. Am. Chem. Soc. 2013, DOI: 10.1021/ja4088874).
The new approach improves on solid-state NMR techniques for determining three-dimensional structures, in large part by sidestepping the need for isotopic labeling of samples.
The team was led by Lyndon Emsley, a chemist at France’s University of Lyon and scientific director of the Lyon-based European Center for High-Field NMR, and Graeme M. Day, a chemist at the University of Southampton, in England.
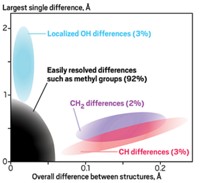
Their technique includes three parts. First, they use computational analysis to develop a set of possible crystal structures of a molecule based on its chemical structure. Then they calculate the 1H NMR chemical shifts each crystal structure would show. Finally, they match the calculated chemical shifts to the experimental spectrum obtained from the powder.
The team reporting the new work analyzed an AstraZeneca drug candidate for type 2 diabetes called AZD8329. The drug has seven known crystal forms; AstraZeneca chose two for development on the basis of their material properties. One of the two, “form 4,” had not been structurally characterized. The Emsley-Day team used its new approach to identify 34 crystal structure candidates computationally—one of the structures exhibited a calculated spectrum that matched that of the form 4 powder.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter