Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Gene Silencing By Design
Drug Discovery: Tweaking short interfering RNAs improves their ability to turn off bad genes
by Stu Borman
November 18, 2013
| A version of this story appeared in
Volume 91, Issue 46

When new protein structures are reported, it’s often claimed they could lead to the design of therapeutic agents that interact with the proteins more effectively. A new study on short interfering RNAs (siRNAs) actually demonstrates such a result.
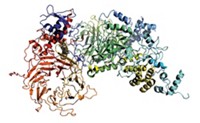
Last year, two research groups determined crystal structures of the human version of Argonaute—a protein in the RNA interference (RNAi) pathway that cells use to turn off target genes. siRNAs activate RNAi in cells by binding to messenger RNAs for targeted genes and marking them for destruction by Argonaute.
siRNAs could thus be used therapeutically to turn off problematic genes related to various diseases. But designing siRNA drugs isn’t easy because they don’t readily enter cells, are broken down and excreted quickly, produce immune reactions, and cause side effects by inhibiting off-target genes. Nevertheless, several chemically modified siRNA drug candidates have entered human clinical trials.
Dean J. Tantillo, Peter A. Beal, and coworkers at the University of California, Davis, have now modified siRNAs in a new way—by using Argonaute’s crystal structures for the first time as a jumping-off point for structure-based siRNA redesign (J. Am. Chem. Soc. 2013, DOI: 10.1021/ja4079754).
siRNAs must bind Argonaute to initiate gene silencing, and the way the siRNAs bind affects potency. The UC Davis researchers used target docking simulations and their own chemical intuition to find siRNA modifications that improve potency by changing the way siRNAs bind Argonaute. Higher potency enables lower doses of a drug treatment, which tends to reduce side effects.
The group’s best redesigned siRNA—in which the terminal nucleotide is replaced with an analog—is nearly 20% more potent than the natural version in lab tests. The increase is only modest but improvable, the researchers believe. “New analogs discovered using our approach that enhance RNAi activity will be patented,” Beal says.
The work should inspire chemical modifications “that address some of the shortcomings of siRNA as potential therapeutics,” comments siRNA-modification expert Masad J. Damha of McGill University.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter