Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Watching Enzymes Change Molecules’ Chirality
Biochemistry: NMR method allows researchers to observe a protein convert a molecule into its mirror image
by Sarah Webb
April 15, 2013

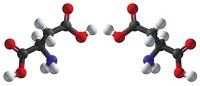
Some antibiotics kill bacteria by blocking the enzymes that the microbes use to build their cell walls. In certain types of bacteria, cell wall construction relies on an enzyme called alanine racemase, which converts the amino acid
Currently researchers study these enzymes by mixing them with one isomer, or enantiomer, and then measuring how much of the mirror-image molecule appears over time using a technique such as circular dichroism. The chemists must take samples from the reaction at certain time points and then analyze them.
NMR can follow the progress of reactions in real time, but the method, on its own, cannot distinguish one enantiomer from its mirror image. Philippe Lesot of the University of Paris-South, in Orsay, and his colleagues previously reported a way to discriminate between the
When she heard about Lesot’s work at a conference, chemist Monique Chan-Huot, a postdoctoral researcher at the Ecole Normale Supérieure, in Paris, wondered if the technique might be useful for studying alanine racemase. She and Lesot soon began working together to adapt Lesot’s technique to observing the enzyme in action. Their main question was whether the enzyme would retain its activity in this unusual liquid crystal environment.
The researchers found that as long as they mixed their samples using a centrifuge so that the enzyme was distributed evenly within the chiral liquid crystals, the conversion rates matched those measured by other techniques. The team next plans to use this NMR technique to study inhibitors of the enzyme as leads for potential new antibiotics.
The method is “a significant new application” of the use of chiral liquid crystals in NMR, says Stefan Berger of the University of Leipzig, in Germany. Chan-Huot and her colleagues are interested in extending this method to other enzymes that react with chiral molecules.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter