Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Drug Development
Aiming For HIV’s Weak Spot
Scientists seek ways to block the virus before it can infect a single cell
by Bethany Halford
September 1, 2014
| A version of this story appeared in
Volume 92, Issue 35

As enemies go, the human immunodeficiency virus has proven particularly shrewd at evading attempts to defeat it. Decades into the fight against HIV, statistics on the virus’s toll are still staggering. Worldwide, approximately 35 million people are infected with HIV, according to the World Health Organization. Last year, complications from the virus claimed the lives of 1.5 million people, and there were 2.1 million new HIV infections.
The ancient Chinese military strategist Sun Tzu advised in his treatise “The Art of War” that one key to success in warfare was to know the enemy. To that end, legions of scientists and doctors have set their sights on understanding the precise workings of HIV, hoping that their findings will ultimately lead to better treatments and preventive medicine that will end the virus’s devastating campaign.
One group’s efforts began nearly two decades ago with reconnaissance on a key protein sitting atop HIV’s surface. The goal was to find a weak spot that scientists might target with a small-molecule weapon. Although progress has been slow, today researchers have not only designed compounds specific for a vulnerable area on HIV, they’re now strategizing how best to deploy them.
BEGINNING SURVEILLANCE
In the 1990s, Wayne A. Hendrickson, a structural biologist at Columbia University, Peter D. Kwong, Hendrickson’s graduate student at the time, Joseph G. Sodroski, a virologist at Harvard School of Public Health, and coworkers set out to solve the crystal structure of envelope glycoprotein gp120, a vital component of HIV’s so-called envelope spikes. These protein protuberances on the virus’s outer coat help HIV attack healthy cells.
The envelope spike consists of three gp120s sitting like petals on a protein stalk. The first step in HIV infection occurs when gp120 interacts with a protein called CD4 on the surface of the immune system’s helper T cells. This interaction causes gp120 to reorganize, changing its conformation so that the protein can subsequently interact with another protein, known as the coreceptor, on the T cell’s surface. In some cells this coreceptor is called CCR5, and in others it’s called CXCR4.
The binding of gp120 to CD4 and the coreceptor draws the stalk toward the healthy T cell so that its tip penetrates the cell membrane. The virus’s outer coat then fuses with the cell’s membrane.
Scientists still don’t know precisely how that fusion happens, but they do know that the process creates a pore through which the virus can discharge its capsid, which contains all of the virus’s genetic information. That genetic information gets incorporated into the cell’s nucleus, transforming it from a healthy cell to a host cell for the virus. And once those genes reach the nucleus, there is no getting rid of them.
MAPPING THE TERRAIN
By 1998, “a lot of biochemistry had already been done on gp120, and we knew a good deal about it, but we knew absolutely nothing about what it looked like,” Columbia’s Hendrickson says. Getting a crystal structure of the protein bound to CD4 would give researchers a picture of that first step in the infection process. With this picture, scientists could perhaps figure out a way of gumming up HIV’s machinery—a strategy for stopping the virus before it gets a foothold.
At the time, scientists suspected that three copies of gp120 were present on the HIV envelope spike, but they didn’t know for sure. It was exceedingly difficult to crystallize the whole envelope spike structure, Harvard’s Sodroski tells C&EN, so the team took a reductionist approach. They decided to look at just one gp120 moiety, and they stripped the molecule of its sugar molecules, which were preventing the researchers from getting good crystals. They added components of the CD4 protein and an antibody fragment that helped with crystallization.
“That in itself was a several-year project,” Sodroski says. By 1998, the team published a crystal structure (Nature, DOI: 10.1038/31405). What they saw told them much about how the virus was evading the immune system, Hendrickson says. Although the researchers had removed the sugars, they knew where the molecules would normally be attached and could tell they were forming a shield for the virus. The sugars were the same as those on proteins in our bloodstream, so the immune system didn’t recognize them as invaders and didn’t attack the virus.
The crystal structure also revealed a hydrophobic cavity at gp120’s center. A key phenylalanine residue on CD4 bound within this pocket. Could this, they wondered, be HIV’s weak spot?
GATHERING FORCES

Irwin Chaiken, then a biochemistry professor at the University of Pennsylvania (he has since moved down the road to Drexel University College of Medicine), had been collaborating with Hendrickson and Sodroski on a related project. When he learned that they were in the process of determining the CD4-gp120 structure, he too saw an opportunity to disrupt, or antagonize, this key protein-protein interaction.
“HIV is a wonderful protein machine,” Chaiken says. “It works by a sequence of interactions. As we discover more about how proteins work, we realize that most proteins work in the ways that you see right here. Interactions cause conformational changes that lead to function.”
There’s quite a bit of pessimism surrounding the notion of disrupting protein-protein interactions with small molecules, Chaiken says. The problem, he explains, is that you’ve got two relatively large objects interacting at their surfaces. Getting a small molecule to disturb that interaction, which takes place at multiple points, isn’t trivial.
But Chaiken thought it was worth a try. He approached Hendrickson and Sodroski about applying for a Research Program Project Grant from the National Institutes of Health. These P01 grants, as they are known, are designed to support broadly based, multidisciplinary research with a specific major objective or a basic theme. They usually combine the efforts of several different research teams, each of which is studying a different aspect of the problem.
Seventeen years later, the P01 project is still ongoing. Seven different teams now work together under its aegis with Chaiken serving as the principal investigator. “For me, the wonder of this project is being able to work with a group of people who can attack this problem from diverse perspectives,” he says. “What’s brought us together and what’s kept us together is the ability to do our own science and, at the same time, to interconnect to enhance the science we’re doing.”
The group’s goal, Chaiken continues, is to figure out the best approaches for blocking HIV. “Our mandate isn’t just to make a drug but also to develop a deep understanding of how gp120 works.”
In 2002, Chaiken asked Amos B. Smith III, a University of Pennsylvania chemist who is known for his work in organic synthesis, to join the project. “They came to me because they needed chemistry on small molecules to address some of the things that were coming out of the project’s computational screening program,” Smith explains.
“In that original X-ray structure from 1998, they saw that there was isopropyl alcohol in gp120’s pocket that came from the crystallization process,” Smith continues. “This meant that the pocket had some volume.” Not only did the phenylalanine residue on CD4 fit into the cavity, but there was also enough room for the isopropyl alcohol.
From there, Hendrickson’s group mutated the phenylalanine residue to a cysteine, and Smith’s group made compounds that alkylated the cysteine’s thiol. The researchers later got crystal structures of these alkylated mutants, giving them a chance to chemically probe the cavity. The surprising thing, Smith says, was that even when the alkylating agents were large, the cavity was able to accommodate them, indicating that there was considerable plasticity in gp120.
With those structures in hand, Smith says, the idea was to design a small molecule that would fit into the pocket and block CD4 from binding to the virus in some way. “We tried, and to be honest, we didn’t get very far,” he says.
CHARGING AHEAD AND FALLING BACK
In 2005, a group led by Asim K. Debnath, a chemist at the New York Blood Center, used high-throughput screening to identify a pair of small molecules that prevented HIV from entering T cells (Virology, DOI: 10.1016/j.virol.2005.06.008). The compounds, NBD-556 and NBD-557, were blocking the interaction between CD4 and gp120.
“This was a very important discovery from our perspective,” Smith says. “It was a breakthrough that allowed us to go forward.” Smith’s group synthesized NBD-556 and sent it off to Sodroski’s lab for further study. They also sent it to Ernesto Freire, a biophysicist at Johns Hopkins University who joined the P01 project around the same time Smith did.
It was at this point that the HIV story took an interesting turn. The researchers found that although NBD-556 binds within gp120’s critical cavity where it can prevent binding with CD4, the compound also gives HIV the power to infect cells that don’t have CD4 but that do have one of two coreceptors: CCR5 or CXCR4. In other words, the compound enhanced viral entry.
What the researchers determined was happening was that NBD-556 was binding to gp120 in the same way CD4 does, and in doing so, it triggered the conformational change that allows gp120 to bind to the coreceptor. In essence, it primed the virus (Biochemistry 2006, DOI: 10.1021/bi061193r).
“It was exciting in one way, and of course, it was disappointing in one way,” Debnath recalls. But he agrees with Smith that the discovery turned out to be a breakthrough. “The whole field actually flourished based on this critical information,” Debnath says. “We started thinking, ‘How can we keep the antiviral property of this NBD-556 molecule but design something that won’t induce the conformational changes in gp120 that are observed with CD4 binding? How can we convert this CD4 agonist into an antagonist?’ ”
Chaiken’s P01 group began to explore the same questions. Freire invented a technique to map which amino acids contributed to CD4’s binding to gp120 and which amino acids were responsible for the conformational change that occurs once that binding takes place. The technique combines alanine-scanning mutagenesis with isothermal titration calorimetry.
Alanine-scanning mutagenesis is a standard technique in which researchers mutate amino acids in a protein, converting them one at a time to alanine, which has only a methyl group for a side chain. They then measure the effect this change has on binding. Freire’s group combined this with isothermal titration calorimetry, which measures the characteristic thermodynamic signature from gp120 when it changes conformation.
“With some amino acids, nothing happens. That means those amino acids don’t contribute to binding or elicit the conformational change,” Freire says. “Other amino acids contribute to binding and not to the conformational change, and some contribute to both.” By figuring out where the amino acids that trigger gp120’s conformational change reside in the cavity, “we can avoid them and only target those amino acids that are involved in binding alone,” Freire adds. He calls these different regions “hot spots” (Chem. Biol. Drug Des. 2013, DOI: 10.1111/cbdd.12075).
CHOOSING A WEAPON
Meanwhile, Smith’s group began working with computational chemist Judith M. LaLonde at Bryn Mawr College, in Pennsylvania. The researchers took a harder look at NBD-556, dividing it into three regions. Region one was an aromatic ring thought to bind within gp120’s hydrophobic pocket. The oxalamide that makes up region two appeared to be hydrogen bonding with two backbone carbonyls of residues on opposite sides within the cavity. And region three, an amine, was likely making a critical hydrogen bond with an aspartic acid on gp120.
Smith says that over the course of several years, his group made more than 300 compounds, guided by computational, biophysical, biochemical, and structural data from the P01 collaborators. First, the researchers began tweaking region one. They found that by adding fluorine to the aromatic ring, creating a compound called JRC-II-191, they could boost affinity for gp120.
Advertisement
Changes to region two only led to inactive compounds, so the researchers concluded the oxalamide moiety was critical. Finally, they began adding different amines to region three, hoping to maximize the interaction with the aspartic acid residue on gp120 that’s just outside the cavity. After a good deal of trial and error, they came up with the compound (+)-DMJ-I-228, which has an indane moiety attached to a guanidinium group. Their thinking was that this guanidinium group might mimic an arginine residue on CD4.
This compound turned out to be a good inhibitor of HIV viral entry and didn’t trigger the dreaded conformational change associated with enhanced viral infection. After crystallizing it with gp120, the researchers noted that adding a single carbon between the indane and the guanidinium might help the latter come in closer contact with the critical aspartic acid.
They prepared that compound, (+)-DMJ-II-121, and found that it was 10 times as potent as (+)-DMJ-I-228 at inhibiting HIV viral entry. And it did not elicit the conformational change in gp120. But when they got the crystal structure of this new compound bound to gp120, they were in for a surprise. Instead of binding the aspartic acid hot spot, the guanidinium group had reached out to a different hot spot on gp120, a methionine residue thought to be associated with binding alone. Smith and his former student and postdoc Joel R. Courter recently detailed the evolution of these compounds in Accounts of Chemical Research (2014, DOI: 10.1021/ar4002735).
When asked how he thinks the work has evolved over the past 17 years, Hendrickson wryly replies, “The first word that comes to mind is ‘slowly.’ ” But this recent success has buoyed the group, he adds. “We’re now talking about drug territory. For a long time, we struggled with things that were not anything like that.”
In terms of drug delivery, the researchers first thought of delivering these small molecules via a microbicide that might be applied vaginally or rectally to prevent infection from sexual fluids. A recent study suggests that DMJ compounds used in this way might also complement vaccine approaches. Sodroski’s group showed certain easily elicited antibodies, which on their own are ineffective at preventing HIV infection, could block the virus when combined with the DMJ compounds (J. Virol. 2014, DOI: 10.1128/jvi.00540-14).
In people, Sodroski says, you would vaccinate to elicit the antibodies and then supply the DMJ compounds via some type of slow-release formulation. The ideal prevention, he adds, is really a one-shot vaccine or something that doesn’t require people to be constantly vigilant. But until such a preventive exists, it’s important to consider every option. “I think we’re looking at all kinds of possibilities for prevention because vaccine development has been so frustratingly slow,” Sodroski adds.
BRINGING IN REINFORCEMENTS
Chaiken’s P01 group has not been the only one going after small molecules of this kind. Debnath has also spent the last nine years trying to build molecules that would prevent CD4 binding to gp120 without triggering a conformational change. It has been something of a struggle, he confesses, but “now we are seeing some light.”
Debnath’s team recently developed a small molecule based on what they’ve gleaned from NBD-556 and related compounds over the years. “It has extraordinary breadth of antiviral activity among inhibitors of this class reported so far,” and it does not promote HIV entry to cells, he says. Debnath hopes to publish the work in the next few months.
Targeting gp120 to prevent HIV infection has also been a goal of scientists at Bristol-Myers Squibb. Nicholas A. Meanwell, executive director of discovery chemistry, tells C&EN that the pharmaceutical company started working on the target more than 15 years ago.
While screening molecules from a library of amides for novel mechanisms of action, the scientists came across one compound that turned out to be working at the earliest stages of HIV infection, Meanwell says. Its potency was modest, but it gave the company’s chemists something to start with. They modified its structure until they had a molecule that was quite potent at inhibiting gp120’s interaction with CD4 and also had a good pharmacokinetic profile.
Initially, the group thought their compounds, including lead compound BMS-626529, were binding in the gp120 cavity, but now they think that’s not the case. “It’s been very difficult for us to understand exactly how our molecules bind,” Meanwell says. “We think they bind adjacent to that pocket and interfere with the conformational change.”
Unfortunately, BMS-626529 suffered from poor aqueous solubility. “Once a solution formulation of BMS-626529 hit the gut, the compound would precipitate,” Meanwell laments. And because it was insoluble, it couldn’t get absorbed.
So the chemists synthesized a phosphonooxymethyl prodrug, which they named BMS-663068 (J. Pharm. Sci. 2013, DOI: 10.1002/jps.23476). This compound is soluble in the gut, where it gets converted into BMS-626529 by alkaline phosphatase enzymes. “Everything had to be just right,” Meanwell says. “If the prodrug is cleaved quickly and the active molecule is absorbed slowly, it would precipitate and you wouldn’t get exposure. If it’s cleaved too slowly, then it gets excreted. We were very fortunate that it worked out.”
According to Bristol-Myers Squibb scientists, their prodrug is the first investigational antiretroviral to bind directly to the virus, prevent initial viral attachment to CD4 T cells, and block HIV entry into those cells. The compound recently went through Phase IIb clinical trials, where it performed as well as an existing retroviral treatment. The results suggest that BMS-663068 is potentially as effective as one of the current standards of care and may provide physicians with another weapon against the virus, especially in patients who have failed a prior HIV regimen and need new treatment options.
James A. Hoxie, director of the Penn Center for AIDS Research, says this small-molecule approach offers certain advantages. “These are small molecules, and the smaller they are, the cheaper they are to make, and the easier they are to formulate,” he says. “The fact that these molecules are so small and have this activity I think could play powerfully into their practical use.
“We shouldn’t ever say that we don’t need any more drugs for HIV,” Hoxie adds. “If you’ve got a new way to go after the virus, and you use that new way in combination with drugs that act on different mechanisms, you can get synergistic effects that can be dramatic.”
The Current Arsenal For Fighting HIV
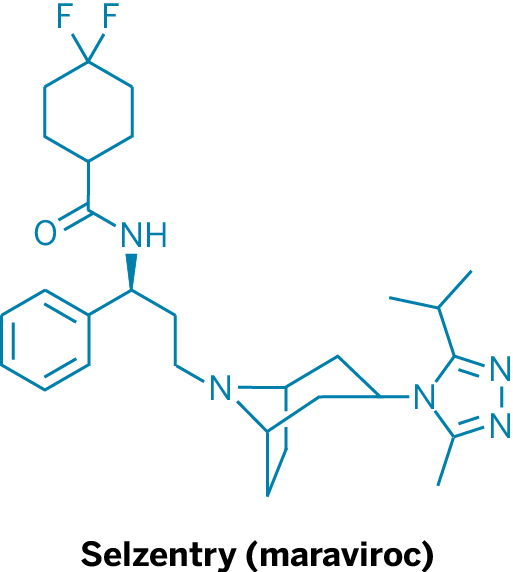
To turn the tide in the fight against HIV, scientists have given doctors many weapons for battling the virus. The most successful of these to date has been HAART, or highly active antiretroviral therapy. This so-called cocktail combines multiple drugs, each working on a different aspect of the HIV replication process. The HAART regimen has been shown to reduce the amount of active virus in patients’ systems, extending life expectancies by decades.
Currently, two drugs on the market target HIV’s entry into T cells. Pfizer’s Selzentry (maraviroc) is a small molecule that binds to the coreceptor CCR5 on T cells, thereby blocking HIV’s gp120 protein from associating with that coreceptor. This protein-protein interaction makes it possible for the virus to penetrate cells. Selzentry is typically used in patients who don’t respond to HAART.
The other approved HIV entry inhibitor is Roche’s Fuzeon (enfuvirtide), a polypeptide that prevents HIV from fusing with healthy cells. It too is used when other therapies fail.
“Even though we have all of these drugs—and we do have lots of them and many are good—the virus is able to develop resistance to every single drug that we have,” notes James A. Hoxie, director of the Penn Center for AIDS Research. “Resistance to drugs is the rule with HIV, but resistance is harder for the virus to acquire when drugs are used in combinations. So, we not only need more drugs, but we need drugs that act in new ways and can be used in new combinations.”





Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter