Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Fluorescence Studies Reveal How Cells Take Out Their Protein Garbage
Molecular Biology: Researchers use single-molecule methods to observe details of the protein degradation process
by Stu Borman
April 9, 2015
| A version of this story appeared in
Volume 93, Issue 15

The breakdown of old, unneeded, and defective proteins in the proteasome, the cellular trash can for proteins, is as essential to cells as garbage pickups are to neighborhoods. A research team now provides a more complete picture of the way cells use peptides to mark proteins for disintegration in the proteasome.
In one type of protein degradation, peptides called ubiquitins are added to proteins to tag them for disposal. The process, which helps keep proteins at proper levels in cells, is of great importance to human health and disease. Its discovery was honored with the 2004 Nobel Prize in Chemistry. But exactly how it works is not completely understood.
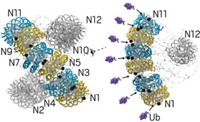
Marc W. Kirschner and coworkers at Harvard Medical School have now used single-molecule fluorescence methods to show, at a fine level of detail, how the enzyme APC/C adds ubiquitins to proteins and how those proteins are then recognized and dispatched in the proteasome (Science 2015, DOI: 10.1126/science.1248737 and 10.1126/science.1250834).
“Both studies substantially enrich our knowledge of ubiquitination and degradation, reveal new properties of APC/C and the proteasome, and challenge established concepts about the ubiquitin-proteasome system,” ubiquitin expert David Komander of the MRC Laboratory of Molecular Biology, in Cambridge, England, notes in a Science commentary.
Using a fluorescence microscope, the researchers observed single molecules of APC/C catalyze the addition of fluorescently labeled ubiquitins to proteins. The team then watched as the proteasome recognized and broke down proteins tagged with different numbers and arrangements of fluorescent ubiquitins.
The experiments showed that APC/C initially adds several ubiquitins to a protein and then uses a process the researchers call “processive affinity amplification” to promote further ubiquitination that directs the protein to its fateful encounter with the proteasome.
They also showed that diubiquitinated proteins are more likely to be degraded in the proteasome than proteins decorated with single ubiquitins or tetraubiquitins. That contradicts a commonly held view in the field that tetraubiquitination is a ticket that proteins always need to be admitted to the proteasome.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter