Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Structural Biology
After Two Decades Of Trying, Scientists Report First Crystal Structure Of A DNAzyme
Structural Biology: Folded conformation with product bound reveals probable mechanism of synthetic catalyst
by Stu Borman
January 7, 2016
| A version of this story appeared in
Volume 94, Issue 2

For about two decades, chemists have been synthesizing libraries of single-stranded DNAs and screening them for ones that can catalyze specific reactions. Scientists think these DNA enzymes, or DNAzymes, could serve as medical diagnostics, functional components in nanotechnologies, or even drugs. In fact, some have been tested in clinical trials, but none have yet been approved.
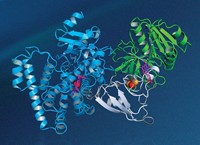
But until now, researchers haven’t been able to obtain a crystal structure or determine a catalytic mechanism for any DNAzyme. By sheer persistence, Claudia Höbartner, Vladimir Pena, and coworkers at the Max Planck Institute for Biophysical Chemistry and Georg-August University found the right conditions to crystallize and structurally analyze a DNAzyme called 9DB1 that stitches together, or ligates, RNA strands (Nature 2016, DOI: 10.1038/nature16471).
They succeeded by crystallizing a form of 9DB1 in which its reaction product, a ligated RNA, is still attached. The structure enabled the researchers to propose the first molecular mechanism of action for any DNAzyme.
The work could help researchers begin to rationally design DNAzymes to improve their catalytic activity or change their substrate selectivity. Indeed, the Höbartner-Pena team revised 9DB1’s substrate selectivity as part of the study.
“It’s the missing piece,” says Gerald F. Joyce of Scripps Research Institute California, who, with Ronald R. Breaker of Yale University, discovered the first DNAzyme in 1994. “I know from personal experience how hard it’s been to get a DNAzyme structure, and this is the first one. It’s really cool to see DNA catalyzing a reaction like this.”
“Many groups have tried to provide a crystal structure of a DNAzyme for the past two decades, and finally one has succeeded,” says functional nucleic acid specialist Yingfu Li of McMaster University. Mutagenesis studies had provided indirect evidence suggesting that single-stranded DNAzymes are capable of adopting intricate tertiary folds like those of enzymes and ribozymes—RNA enzymes—to form well-defined catalytic active sites. But this study is the first to make it crystal clear, Li says.
Scott K. Silverman of the University of Illinois, Urbana-Champaign, who first identified 9DB1 and was Höbartner’s postdoc adviser, notes that the mechanistic analysis reveals how the positioning of key nucleotide pairs in 9DB1 enables it to catalyze 3’ to 5’ RNA ligation rather than 2’ to 5’ ligation.
The study also highlights a difference between ribozymes and DNAzymes. Ribozymes use RNA’s 2’-hydroxyl groups, which are absent in DNA, for structural interactions or directly for catalysis. The new structure shows why the lack of these groups doesn’t diminish the catalytic activity of DNAzymes. The missing hydroxyls make DNA’s sugar-phosphate backbone more flexible, allowing acrobatic conformations that compensate for the absent hydroxyls in DNAzymes.
Höbartner admits that the study could have provided more mechanistic information if the DNAzyme had been caught prior to or during catalysis, as such conformations are more mechanistically revelatory. “Our next goals are to obtain a structure of 9DB1’s precatalytic state and to get further insights into the mechanism,” she says. She and others in the field also hope eventually to structurally analyze other DNAzymes and compare their mechanisms with that of 9DB1.
This article has been translated into Spanish by Divulgame.org and can be found here.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter