Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
ACS Meeting News
Small-molecule selectively destroys cancer-associated RNA
Researchers stop cancer progression and kidney disease in mice using modified, repurposed drug
by Megha Satyanarayana
August 26, 2021
| A version of this story appeared in
Volume 99, Issue 31

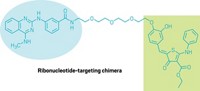
Researchers have converted an existing cancer drug into a molecule that selectively targets a cancer-associated ribonucleic acid (RNA) for destruction. This molecule, a ribonuclease targeting chimera, or RIBOTAC, stops disease progression in mice engineered to have breast cancer.
Matthew Disney, a medicinal chemist at Scripps Research, led the work and presented the findings at the American Chemical Society Fall 2021 meeting last week. He is also the cofounder of Expansion Therapeutics, a start-up developing drugs that target RNA.
While most drugs target proteins, several diseases are associated with the presence or absence of certain RNAs, so targeting RNA could also help treat disease. Disney said that current RNA modulators rely on small strings of nucleotides, which are challenging to get into cells. Replacing those strings with small molecules that bind to certain structures in RNA as it folds and bends could improve a therapy’s performance. “This same technology could apply to cancer or anything,” he said.
Amanda Garner, a chemical biologist at the University of Michigan, said the work is exciting because Disney’s RIBOTAC approach turned a less specific event—a drug binding to a family of RNAs—into something highly specific that degrades a particular target RNA.
Using an existing drug library and an RNA library representing more than 1,000 unique 3D folded structures, Disney and his team found 68 known drugs that could bind to members of the RNA library. Four of those 68 bound RNA particularly strongly; they included dovitinib, a receptor tyrosine kinase (RTK) inhibitor used to treat breast cancer. RTKs are involved in multiple aspects of breast cancer progression.
Looking more closely, the team found that dovitinib binds microRNA-21 (miR-21)—a small bit of RNA that suppresses protein production by blocking the cell’s translation machinery. Overexpression of miR-21 is also associated with cancers.
Since dovitinib preferentially interacts with the RTK inhibitor protein it was designed to inhibit, Disney and his team searched for ways to skew its action to block miR-21. They converted dovitinib to a RIBOTAC by adding a chemical moiety with a repeating carbon and oxygen chain and a five-member ring containing sulfur that could recruit and activate an RNA-degrading enzyme called ribonuclease L (RNase L). This tweak, Disney said, improved the selectivity of dovitinib for the miRNA about 2,500-fold. The team tested its RIBOTAC (shown) in several types of cancer cells and showed that miR-21 levels decreased (J. Am. Chem. Soc. 2021, DOI: 10.1021/jacs.1c02248).
The researchers found that an injection of the RIBOTAC led to fewer cancer nodules in mice engineered to have metastasized breast cancer; it also led to improved kidney function in mice engineered to have a kidney disease that is also associated with miR-21.
Matt Woll , a medicinal chemist and the vice president of chemistry at PTC Therapeutics, said designing an RNA-modifying drug is no small feat. Both Woll and Garner said any additional technology to modulate RNA is welcome.
RNA has traditionally been a difficult drug target, Garner said. It is dynamic—with folds and loops, and difficult to crystallize—which complicates the search for drug candidates by targeting structural elements. RNA can be chemically modified by the cell, she said, and it’s often covered with proteins, hiding possible small-molecule binding sites.
Disney agreed. This study was a test to see if the idea of a RIBOTAC would even work, he said. RNA does have some stable folds, but its overall instability means there aren’t always clear and expected places where drugs can bind. “You don’t know what’s the best target until you have it,” he said.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter