Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Structural Biology
Covid-19
Characterizing SARS-CoV-2 antibodies
Neutralizing antibodies for the new coronavirus can be classified into 4 distinct groups to help treatment design, researchers say
by Laura Howes
October 15, 2020
| A version of this story appeared in
Volume 98, Issue 40

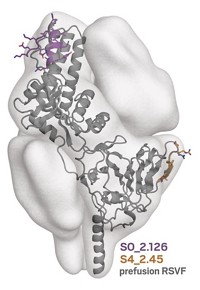
Antibody therapeutics will be a key part of the fight against COVID-19, but some do a better job of neutralizing SARS-CoV-2 than others. These antibodies stop infection by blocking the virus from entering cells. Researchers have now used structural biology techniques and binding assays to show four basic modes by which different neutralizing antibodies bind to the novel coronavirus’s spike protein (Nature 2020, DOI: 10.1038/s41586-020-2852-1).
Support nonprofit science journalism
C&EN has made this story and all of its coverage of the coronavirus epidemic freely available during the outbreak to keep the public informed. To support us:
Donate Join Subscribe
These antibodies specifically target the protein’s receptor-binding domain, or RBD. “There was evidence of multiple binding modes, or targets, on the RBD surface,” explains Christopher O. Barnes, a postdoc in Pamela Bjorkman’s biology lab at the California Institute of Technology. To explore these domains, Barnes used cryo-electron microscopy to visualize spike-protein-bound neutralizing antibodies isolated from people with COVID-19. He could see not just where antibodies might bind to one RBD but also how the antibodies might bind to the full spike and how it might be possible for different antibodies to double up on one spike.The spike is a complex of three identical proteins, each of which has its own RBD. The Caltech team classified four complementary ways in which the neutralizing antibodies bind to different sites and orientations of these RBDs.
Many pharma firms are working on getting antibodies for SARS-CoV-2 into the clinic. A promising approach is to use a combination of them. The Regeneron antibody mix that President Donald J. Trump took as part of his treatment for COVID-19 consists of two individual antibodies. Those two antibodies correspond to potentially complementary classes, according to Bjorkman and Barnes’s system.Michael Hust, who is developing SARS-CoV-2 neutralizing antibodies at the Technical University of Braunschweig, cautions that there are also differences within the classes. “It is not black and white,” Hust says.
Bjorkman is working with a group using computational techniques to optimize the binding of the antibodies. But Bjorkman says she’s not sure if that will be needed—the ones some patients have made naturally are quite effective. While we still have much to learn about this virus and our immune response to it, Bjorkman hopes the work can help researchers predict what antibodies might perform better and why, and which cocktails might prove most effective at fighting the pandemic.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter