Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Structural Biology
Enzymes with opposite functions form complex
Bacterial enzyme pair regulates glutamate by inhibiting one of the enzymes
by Celia Henry Arnaud
January 5, 2022
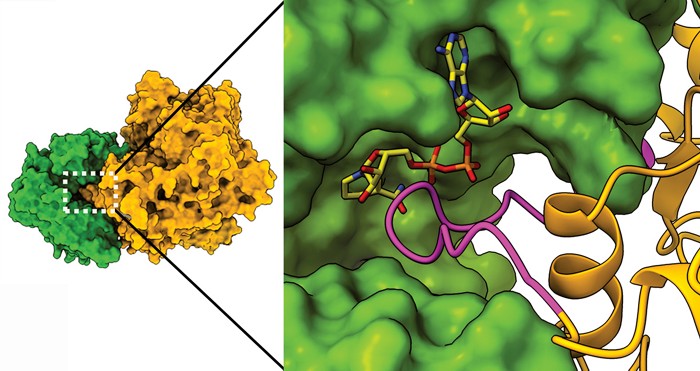
Usually enzyme complexes make multistep processes easier by catalyzing sequential reactions in which the product of one enzyme becomes the substrate for the next. In a new finding, researchers report an enzyme complex where the two interacting enzymes have opposite functions—one makes the metabolite glutamate and the other degrades it. When bound to each other, one of the enzymes inhibits the activity of the other. In this way, the complex has the effect of regulating the amount of the glutamate inside the bacterial cell.
Researchers led by James S. Fraser of the University of California San Francisco and Dan S. Tawfik and Vijay Jayaraman of the Weizmann Institute of Science solved cryo-electron microscopy structures of a complex of the enzymes glutamate synthase (GltAB) and glutamate dehydrogenase (GudB) in Bacillus subtilis. The structures reveal that the two enzymes form a complex in which one of GltAB’s subunits blocks the active site of GudB. GltAB catalyzes glutamate formation, but when there’s too much glutamate present, the cell shuts down GltAB but not GudB production. Without GltAB to block it, GudB can degrade the excess glutamate (Nat. Chem. Biol. 2021, DOI: 10.1038/s41589-021-00919-y).
Hints of such complexes have been seen before, Fraser says. “This is the first time that we’ve ever seen the structural basis of how this occurs,” he says.
Alisdair Fernie, who studies metabolism at the Max Planck Institute of Molecular Plant Physiology, suspects that other similar regulatory complexes will be found. “Our eyes are focused on these assemblies optimizing efficiency so much so that we were blinded from seeing the opposite.”


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter