Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Structural Biology
Following a code for swapping amino acids makes membrane proteins water soluble
Detergent-free versions could facilitate biological studies
by Celia Henry Arnaud
August 29, 2018
| A version of this story appeared in
Volume 96, Issue 35
Membrane proteins play important roles in health and disease, but they are so hydrophobic that they require detergents to maintain their shape outside membranes. That makes them difficult to study. But what if those proteins could be tweaked to be hydrophilic while maintaining their usual structure and function?
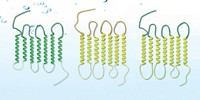
Shuguang Zhang of Massachusetts Institute of Technology and coworkers have figured out a way to do just that. By swapping hydrophobic amino acids for structurally similar hydrophilic ones, the researchers transform hydrophobic transmembrane receptors into water-soluble proteins that don’t need stabilizing detergents (Proc. Natl. Acad. Sci. USA 2018, DOI: 10.1073/pnas.1811031115).
To achieve these changes, the researchers used what they call the QTY code. They replace leucine with glutamine, which has the one-letter abbreviation Q; isoleucine and valine with threonine (T); and phenylalanine with tyrosine (Y). The substituted amino acids have electron densities similar to those of the amino acids they replace. And because they are neutral, they shouldn’t alter the properties of the proteins.
As a case study the researchers used four chemokine receptors, G protein-coupled receptors with seven transmembrane domains. These receptors are among the most intensely studied drug targets. Three of them are coreceptors for HIV entry into T cells. Crystal structures have previously been solved for two of them.
At first, Zhang and coworkers replaced amino acids only on the protein surface. But the proteins still needed detergents. Then they replaced more, but it still wasn’t enough. “We finally replaced all the leucine, isoleucine, valine, and phenylalanine in the transmembrane domains,” Zhang says. “Then the QTY-code-designed proteins became detergent-free and water soluble.”
They’re currently working to obtain crystal structures of the modified proteins. But in molecular simulations, the structures of the hydrophilic proteins line up well with crystal structures of the original receptors.
“Even in the absence of a more detailed computational characterization or actual experimental structures, this work has all the ingredients of a landmark paper,” says Martin Egli, a structural biologist at Vanderbilt University. “The discovery of a simple code to convert a very large number of hydrophobic to hydrophilic amino acids in the membrane-spanning region of chemokine receptors for successful solubilization in water, without apparently impacting fold and activity in a dramatic fashion, raises the prospect of extending this approach to other classes of transmembrane proteins, including transporters and ion channels.”
And that’s what Zhang’s team plans to do next. “Although we have not done experiments for other proteins, we believe the QTY code could be widely applicable to other aggregated and water-insoluble proteins,” Zhang says.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter