Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Structural Biology
Covid-19
Structure of SARS-CoV-2 RNA-dependent RNA polymerase published
Researchers use cryo-EM to image the viral protein that remdesivir targets
by Laura Howes
April 15, 2020
| A version of this story appeared in
Volume 98, Issue 15

Researchers studying SARS-CoV-2, the novel coronavirus that causes COVID-19, have published another structure of a viral protein that could be a target for treatments, including Gilead Sciences’s remdesivir. A team in China led by Zihe Rao at Tsinghua University report the cryo-electron microscopy (cryo-EM) structure of the RNA-dependent RNA polymerase (RdRp), which is a key part of the virus’s replication machinery that makes copies of its RNA genome (Science 2020 10.1126/science.abb7498).
RdRp is one of the main drug targets for SARS-CoV-2, along with the virus’s spike protein, which is involved in infecting human calls, and its main protease, which helps process viral protein production. Jason McLellan, the University of Texas at Austin researcher who published the first cryo-EM structure of the spike protein last month, says RdRp is the only other protein that he was interested in investigating. But before he could start work on the protein, he saw that Rao's group had successfully solved the structure and uploaded a preprint of its work to bioRxiv. He points out that Rao’s group also recently solved the structure of the coronavirus protease. These studies are key, he says, “to give you a clue of what you need and then also how to optimize it.”
Support nonprofit science journalism
C&EN has made this story and all of its coverage of the coronavirus epidemic freely available during the outbreak to keep the public informed. To support us:
Donate Join Subscribe
The novel coronavirus’s RdRp exists as a complex, with the polymerase, called nsp12, bound with some smaller proteins known as nsp7 and nsp8. Like other RdRps, this one adopts a shape that looks like a right hand with so-called fingers, thumb, and palm domains.
In May 2019, a different group of researchers used cryo-EM to solve the structure of the RdRp for SARS-CoV, the virus responsible for the severe acute respiratory syndrome (SARS) outbreak in 2002-3 (Nat. Commun 2019, DOI: 10.1038/s41467-019-10280-3). That virus is similar to the one causing COVID-19. The structure of the SARS-CoV-2 polymerase is at a slightly higher resolution than the SARS-CoV structure and adds some more detail, but the structures are mostly the same. The two polymerases have near identical amino-acid sequences and the active site that catalyzes RNA polymerization is identical.
A key advantage of targeting the SARS-CoV-2 polymerase for drug development, says Sarah Butcher at the University of Helsinki, is that drugmakers have experience with designing polymerase inhibitors for HIV therapies. “There’ve been huge developments in polymerase inhibitors in the past few years,” she says, and the way to gum up a polymerase is conserved across most RNA-based viruses.
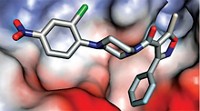
One of the drugs that could inhibit the SARS-CoV-2 RdRp is remdesivir, an antiviral originally developed by Gilead to target the Ebola virus. Phase III clinical trials have started to see if this drug could treat COVID-19 in patients and the first pieces of data from those trials are starting to be published.
Rao and colleagues used their new structure and the structures of similar enzymes to model how the triphosphate form of remdesivir binds to the polymerase. The team thinks that another polymerase-targeting drug, favipiravir, might bind in the same way, suggesting that this could be a general drug target for medicinal chemists designing new antiviral therapies.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter