Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Cell wall model proposed
March 13, 2006
| A version of this story appeared in
Volume 84, Issue 11
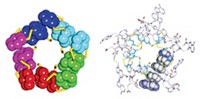
The structure of the bacterial cell wall, which is a target for antibacterial drugs, has been difficult to clarify because of its complexity and the difficulty in obtaining individual components. The cell wall is formed from a peptidoglycan scaffold consisting of repeating disaccharide units of N-acetylglucosamine (NAG) and N-acetylmuramic acid (NAM), with a pentapeptide attached to each NAM. University of Notre Dame chemistry professor Shahriar Mobashery and coworkers have obtained a 3-D NMR structure of a 2-kilodalton synthetic fragment of this scaffold (Proc. Natl. Acad. Sci. USA, published online March 9, dx.doi.org/10.1073/pnas.0510182103). From this fragment, they predict that the longer native peptidoglycan strand is a right-handed helix with three repeats per turn and threefold symmetry along the helical axis; each strand can cross-link with up to three neighboring strands. They extrapolate to a model of the bacterial cell wall with a layered honeycomb pattern of varying pore sizes (top view shown), depending on the number of cross-links. Waldemar Vollmer of the University of Tübingen in Germany cautions that the model is "highly speculative" and "contradicts a number of experimental data."



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter