Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Dicer Yields Its Structural Secrets
Architecture of RNAi enzyme suggests it measures and then snips its substrates
by Stu Borman
January 16, 2006
| A version of this story appeared in
Volume 84, Issue 3

The first crystal structure of Dicer, an enzyme that initiates RNA interference (RNAi), suggests how it works. RNAi is a process in which short RNA molecules recognize messenger RNAs and mark them for destruction, blocking protein synthesis and gene expression. Interest in RNAi as a tool to study cancer, development, and stem-cell differentiation and as a mechanism for RNA-based therapeutics has skyrocketed in recent years.
The Dicer structure was obtained by a team led by Jennifer A. Doudna, a professor of molecular and cell biology at the University of California, Berkeley, and a Howard Hughes Medical Institute investigator (Science 2006, 311, 195).
"It's absolutely fantastic work," says Alexander Wlodawer, chief of the Macromolecular Crystallography Laboratory at the National Cancer Institute, Frederick, Md. "RNAi is incredibly hot these days, and we all wanted to know how an enzyme that does it looks."
Dicer's role in RNAi is to cleave double-stranded RNAs into fragments called short interfering RNAs (siRNAs) or microRNAs (miRNAs). Single strands from the siRNAs or miRNAs then get incorporated into a multiprotein complex called RISC (RNA-induced silencing complex), which interacts with target mRNAs to inhibit gene expression.
Molecular mechanisms by which Dicer processes double-stranded RNA into siRNAs and miRNAs had been proposed by several groups but could not be confirmed because Dicer's structure was unknown. "Our lab and others worked on human Dicer, and it was no dice," Wlodawer says. "Nobody managed to get good crystals."
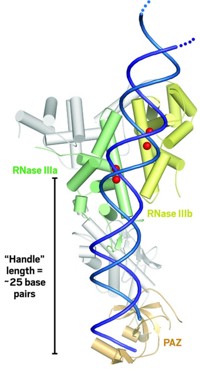
Doudna and coworkers were able to induce a smaller version of Dicer found in protozoa to form analyzable crystals. They then determined the protein's structure by using strong X-rays from a synchrotron. The 3.3-Å-resolution structure shows Dicer to be shaped like a hatchet-appropriately enough, considering its role in RNA cleavage. Two ribonuclease III (RNase III) catalytic domains form the blade, an extended α-helix is the handle, and an RNA-binding module called PAZ forms the base of the handle.
The enzyme's RNA substrate most likely lines up along the handle. The distance between PAZ and the RNase III domains is about 25 RNA base pairs long, the same length as Dicer's RNA fragment products. So the presumption is that Dicer acts like a molecular ruler by cutting its RNA substrates into pieces whose size depends on the length of the enzyme's handle. The researchers also propose that two metal ions in each RNase III domain participate in the mechanism of the enzyme's catalytic action.

The new structure is "a first step because it is still fairly low resolution and this small Dicer does not have all the domains found in its human version, but a number of the group's findings are beyond any doubt, such as the two-metal-ion mechanism," Wlodawer says. "So the structure already tells us a lot, and the work will certainly be extended at some stage."
"It is an important breakthrough, and it certainly will stimulate a lot of research in the area," says RNAi specialist Witold Filipowicz of Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland. "What is missing is that one would like to have a cocrystal" of Dicer with its RNA substrate to prove the mechanistic model. Filipowicz notes that Xinhua Ji, a colleague of Wlodawer's at NCI Frederick, is currently close to determining the structure of RNase III bound to its RNA substrate and that this work will complement the Dicer structure.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter