Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
NMR Analysis Of Larger Biomolecules
Technique reveals motions of key residues in huge proteasome complex
by Stu Borman
January 25, 2007

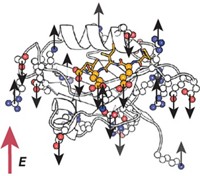
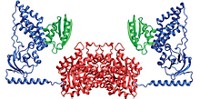
A new nuclear magnetic resonance technique has made it possible to obtain information on the dynamics of key amino acid side chains in the proteasome, a huge protein complex much larger than other biomolecules generally analyzed by NMR (Nature, DOI: 10.1038/nature05512).
The technique, developed by postdoc Remco Sprangers and biochemistry professor Lewis E. Kay of the University of Toronto, has two aspects. One is an isotope-labeling scheme that involves protonation of methyl groups in leucine, isoleucine, and valine residues in a protein that is otherwise highly deuterated. This scheme simplifies the spectrum by permitting the NMR spectrometer to focus only on the labeled methyl groups and essentially ignore everything else.
This isotope-labeling scheme is combined with transverse relaxation optimized spectroscopy (TROSY), an approach that optimizes spectral sensitivity and resolution by increasing the time over which NMR signals from methyl groups can be detected. A related TROSY method was developed earlier by biophysics professor Kurt Wüthrich and coworkers at the Swiss Federal Institute of Technology, Zurich.
The new technique makes it "possible to overcome the molecular-weight limitations that have traditionally hampered quantitative NMR spectroscopy studies" of large biomolecules, such as the 670-kilodalton proteasome, Sprangers and Kay write. The use of NMR to determine high-resolution structures of biomolecules at atomic detail generally remains restricted to much smaller systems, typically having masses of 50 kDa or less. For example, stereo-array isotope labeling (SAIL), a technique developed by chemistry professor Masatsune Kainosho of Tokyo Metropolitan University and coworkers, made it possible for them to obtain the high-resolution structure of the 41-kDa maltodextrin-binding protein (Nature 2006, 440, 52).
The new technique is not a high-resolution structural approach. Instead, the NMR analyses carried out by Sprangers and Kay analyze millisecond, nanosecond, and picosecond dynamics of methyl groups, which tend to move around a lot when the proteasome does its job of catalyzing protein degradation.
Sprangers says: "We found that the walls of the chambers of the proteasome are very mobile. This mobility can be important for translocating substrate proteins from the outside world toward the proteasome's active sites," which are located deep in the interior of the barrel-like structure.
"The approach allows us to look at supramolecular machines and get quantitative information. It sets up a framework for being able to address really neat questions about how these machines work and the role of dynamics in their function," Kay adds.
"This is an absolutely stunning piece of work," says Ad Bax, chief of biophysical NMR spectroscopy at the National Institutes of Health's Laboratory of Chemical Physics in Bethesda, Md. He notes that the Kay lab developed clever new methods for obtaining sharp resonances for methyl groups in large protein complexes such as the proteasome and figured out how to attribute each resonance to a specific methyl group. "They also show that quantitative measures for the rates at which these groups move around within the proteasome particle can be obtained by NMR," Box notes.
The ability to determine the structure of molecular complexes as large as the proteasome "has been considered to be the exclusive realm of X-ray crystallography until now," says Michele Vendruscolo of the University of Cambridge. "This work shows that NMR spectroscopy represents a uniquely powerful way to obtain detailed knowledge about the dynamics of such complexes, information that is not readily accessible through crystallographic studies. Since dynamics are often crucial for function, as illustrated for the proteasome by Sprangers and Kay, NMR is poised to play an indispensable role in structural and cell biology."
NMR spectroscopist Dennis Torchia of NIH's National Institute of Dental & Craniofacial Research adds that the new technique should be useful for obtaining information about internal biomolecular dynamics and interactions, not only in the proteasome but in a wide range of other molecular machines as well. By combining this information with structural data, researchers will be able to achieve a deeper understanding than is currently possible of the way these machines work on a molecular level, he says.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter