Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Protein-Binding Domains Defy Pigeonholing
Model predicts binding partners of domains that mediate crucial protein-protein interactions
by Stu Borman
July 24, 2007
A new study challenges the conventional wisdom about the way in which a key family of protein domains that mediate protein???protein interactions are classified. And it offers a way to predict these domains' preferred protein binding partners.
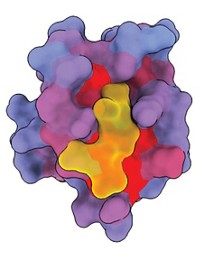
Protein sequences that mediate protein—protein interactions by binding selectively to the ends of target proteins are known as PDZ domains. These domains have traditionally been pigeonholed into different classes according to their binding preferences.
Gavin MacBeath and coworkers at Harvard University have now found that this way of assigning PDZ domains is not justified. "Instead, PDZ domains lie on a selectivity continuum," MacBeath notes. "It appears that PDZ domain binding selectivity is optimized across the proteome to minimize cross-reactivity," the binding of PDZ domains with multiple targets.
The surprising result came from a comprehensive study of 157 mouse PDZ domains—most of the known PDZ domains in the mouse genome—using protein microarrays and fluorescence detection (Science 2007, 317, 364). On the basis of the results, MacBeath and coworkers created a model that can be used to predict the target proteins to which different PDZ domains are likely to bind. The information could be useful to researchers who study PDZ-mediated processes, such as directing the specificity of cell signaling and directing protein trafficking.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter