Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Microspheres Read DNA Microarrays
Electrostatic repulsion detects DNA pairing
July 2, 2008

DNA microarrays—devices that measure gene expression—could become easier to use, thanks to a new readout method based on electrostatic repulsion. Such devices are used in biological research and clinical diagnostics.
University of California, Berkeley, chemistry professor Jay T. Groves, postdoc Khalid Salaita, and graduate student Nathan G. Clack have devised a way to use negatively charged silica microspheres to detect spots on DNA microarrays where single-stranded DNA hybridizes to form double-stranded DNA (Nat. Biotechnol. DOI: 10.1038/nbt1416).
"We're throwing millions of particles on the sample and letting the particles scan the surface randomly by Brownian motion," Groves says. "By doing a statistical analysis of their motion, we're able to determine with high precision what the charge on the surface is."
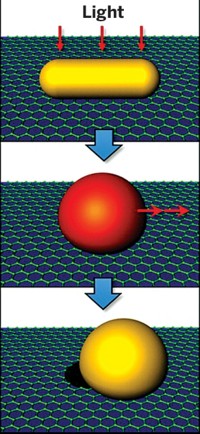
The setup consists of a glass surface modified with aminosilanes so that it has a positive charge. The researchers tune the charge such that single-stranded DNA printed on the surface makes the net charge approximately neutral or only slightly positive. Any DNA added to the surface as a result of hybridization drives the overall charge toward negative at that spot. When a solution of the negatively charged microspheres flows over the surface, the beads move away from the surface at the negatively charged spots where double-stranded DNA is present.
The researchers monitor the position of the spheres with optical methods such as reflection interference contrast microscopy or darkfield or brightfield microscopy. Such detection methods may not always be necessary, however, because the microspheres scatter light so strongly that the pattern of spots with hybridized DNA can be seen with the unaided eye, Groves says.
"The way to think of it is like a dust for fingerprints, except you can dust for DNA hybridization," Groves says. "You could imagine having a glass chip out in the field in a developing country, doing a tuberculosis DNA screen to see which strain you have, and just holding it up to the light to see."
Using a "charge ladder," in which a series of neighboring spots with the same DNA sequence have slightly different charges, the visual method becomes quantitative. "As you go up this ladder, it takes progressively more hybridization to get the spot to switch from positive to negative," Groves says. The last spot in the ladder to switch reveals how much DNA has hybridized.
The Berkeley team also used the method to detect RNA taken from cells without amplification or labeling. "We imagine this taking what right now is a difficult outsourced problem of doing gene expression profiling and turning it into something that you can do quickly in the lab," Groves says. Groves hopes that the technology will be commercialized. "We hope to see a company help develop the technology for real-world applications," he says.




Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter