Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
RNA-Controlled Gene Expression
Target of transcription-altering synthetic RNA is another RNA
by Celia Henry Arnaud
July 10, 2008

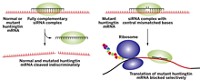
Researchers have identified the target of synthetic RNAs that can turn gene transcription on and off. The finding could lead to new ways of controlling gene expression in cells.

The synthetic RNAs with transcription-altering ability, also known as antigene RNAs (agRNAs), are short, lab-made pieces of double-stranded RNA that are complementary to regions of genomic DNA that promote gene transcription. They inhibit or activate gene expression, depending on the circumstances (C&EN, Feb. 5, 2007, page 8, and Aug. 8, 2005, page 10). But their target was unknown.
Now, the same team that first synthesized agRNAs, led by David R. Corey and Bethany A. Janowski of the departments of pharmacology and biochemistry at the University of Texas Southwestern Medical Center, Dallas, shows that agRNAs bind to noncoding RNA transcripts that overlap the promoter regions of DNA (Nat. Struct. Mol. Biol., DOI: 10.1038/nsmb.1444). A noncoding RNA transcript is single-stranded RNA that does not code for protein.
In subsequent steps of gene activation or inhibition, the agRNA recruits Argonaute proteins (catalytic RNA-binding proteins) to the noncoding transcript, and the resulting complex acts as a scaffold for other transcription factors.
"This paper represents another big step forward in the exciting, rapidly evolving field of RNA-controlled gene expression," says Anna K. Mapp, an associate professor of chemistry and medicinal chemistry at the University of Michigan who studies the regulation of gene transcription. "Janowski and Corey have addressed a key question regarding the function of agRNAs as upregulators and downregulators of transcription by identifying a molecular target."
Using the progesterone receptor as a model system, Corey and Janowski's team shows that agRNAs activate or inhibit gene expression in different circumstances. In a breast cancer cell line that expresses high levels of the receptor, agRNAs inhibit gene expression. In another breast cancer cell line that expresses low levels of the progesterone receptor, the agRNAs activate gene expression. In both cases, noncoding RNA transcripts must be present for agRNAs to work.
Corey points out that such activity is reminiscent of hormone-mediated gene regulation, in which small-molecule ligands can increase or decrease expression levels. "The fact that you can get activation and repression in different circumstances may be something that is more likely to happen in genes that can be easily turned on and off," he says.
However, he expects this mechanism will not be limited to one type of gene. "We've done computational searches of promoter sequences against known microRNAs," Corey says. MicroRNAs are small endogenous RNAs that are complementary to messenger RNAs. They are similar in length to agRNAs and are found throughout the genome. Some of those microRNAs may turn out to act as agRNAs. "The number of excellent matches that we're getting computationally with gene promoters and microRNAs is going up very quickly. There are a lot of candidate genes out there," he says.
The next step is to find endogenous agRNAs, Corey says. "The most important thing is to establish with a high degree of evidence that there are natural agRNAs," he says. "There's a fair amount of skepticism about it. We accept that, and we're going to set a high bar for this work."


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter