Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Physical Chemistry
Gamers Solve Protein Structure
Foldit players find model that leads to solution of retroviral protease structure
by Celia Henry Arnaud
September 26, 2011
| A version of this story appeared in
Volume 89, Issue 39
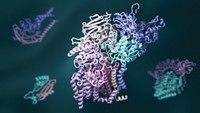
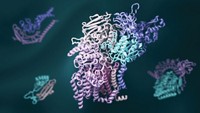
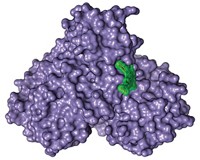
Players of the online computer game Foldit have succeeded in solving a retroviral protease crystal structure that had eluded scientists for years (Nat. Struct. Mol. Biol., DOI: 10.1038/nsmb.2119). Foldit grew out of the distributed computing program Rosetta@home, run by David Baker of the University of Washington, Seattle, in which individuals donate computing power to solve problems in protein folding and design. Foldit players—alone or in teams—compete to find a protein’s lowest energy state within the allotted time. With many proteins, Foldit players run into the same problems that plague other computational methods, getting trapped by local minima in the energy landscape. But Foldit also shows great promise. For example, players were asked to optimize the crystal structure of Mason-Pfizer monkey virus (M-PMV) retroviral protease. Despite having X-ray diffraction data, scientists have been unable to solve the structure because they lacked appropriate crystallographic phase information. Starting with 10 poorly scoring NMR models, one of the teams, the Foldit Contenders group, built on each other’s models, ultimately producing one that solved the phase problem. Within days, scientists had the final structure.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter