Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Analytical Chemistry
Method Spots DNA Modifications On Single Strands
Epigenetics: Patterning approach may help researchers monitor DNA methylation in individual cell
by Christine Herman
October 25, 2011

A new single-molecule technique can spot methylated bases onsingle DNA fragments (Anal. Chem., DOI: 10.1021/ac202506j). The method could help researchers and doctors understand at the level of individual cells how these so-called epigenetic modifications regulate gene expression and lead to disease, the researchers say.
Methylation of the DNA base cytosine can turn genes off and researchers suspect that this modification plays a role in diseases, such as diabetes, Alzheimer’s, and cancer. Traditional techniques for detecting DNA methylation involve either grabbing methylated DNA fragments with antibodies or amplifying methylated DNA fragments using the polymerase chain reaction. These techniques mix DNA from hundreds or thousands of cells and report the average level of methylation.
But sometimes researchers and doctors want to understand how methylation levels differ within a collection of cells, such as in a tumor biopsy. Unfortunately, existing techniques dilute the signals of a few rare, yet critically important diseased cells, says Harold G. Craighead of Cornell University. If scientists could look at single DNA molecules, they could then spot sub-populations of cells that have different methylation patterns from the rest of the population.
To develop a technique for analyzing DNA methylation in a single cell, Craighead and his team adapted a technique previously developed by his postdoc, Aline Cerf, for generating arrays of single DNA molecules (J. Mater. Res., DOI: 10.1557/jmr.2010.12). In the new method, the researchers incubate solutions of DNA fragments with two fluorescent labels: a dye that labels DNA and a fluorescently labeled peptide that binds to methylated cytosines.
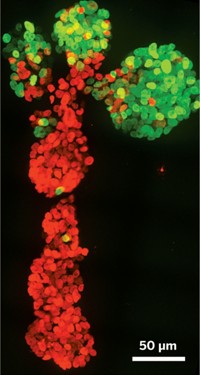
They inject the solution between a glass coverslip and a silicone surface that has rows of evenly spaced 5-µm-deep wells. The scientists then move the glass coverslip horizontally at a constant velocity, while keeping the silicone surface stationary. This process drags the liquid meniscus of the solution across the silicone surface. When a DNA fragment falls into a well as the meniscus crosses it, one end of the strand gets caught. Meanwhile, the moving meniscus pulls the other end of the DNA along the surface. In the end, DNA fragments sit on the surface as straight, elongated, single strands.
The researchers then transfer the stretched-out strands to another glass coverslip, keeping their pattern intact, for imaging by fluorescence microscopy. The two dyes allow the team to spot all of the DNA strands on the coverslip and the regions of each strand that have methylated cytosines.
The researchers tested their method with mixtures of methylated and non-methylated DNAs. They demonstrated that they could selectively detect the methylated strands.
The team hopes eventually to couple this patterning approach with high-resolution imaging techniques to reveal the precise location of epigenetic modifications within DNA molecules isolated from single cells, Craighead says. Researchers in Craighead’s lab have already applied the DNA patterning approach to generate arrays of single DNA molecules on graphene for analysis with electron microscopy (Nano Lett., DOI: 10.1021/nl202219w).
Robert Riehn, a physicist from North Carolina State University, calls the method innovative. Other research groups have used microfluidics to stretch out single DNA strands, he says, but by producing a regular array, Craighead’s microwell method gives researchers more control over separating out single molecules.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter