Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Pharmaceuticals
Prostate Cancer Target Analyzed
Molecular Structure: Detailed view of cancer-related cytochrome P450 could aid drug design
by Stu Borman
January 30, 2012
| A version of this story appeared in
Volume 90, Issue 5

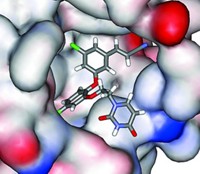
To understand better how two new anticancer agents work, researchers have obtained the first X-ray structures of a key cytochrome P450 enzyme to which they bind. Understanding how the drugs inhibit the enzyme could aid the design of more effective medications for prostate and breast cancer with fewer side effects.
Cytochrome P450 17A1 (CYP17A1) is found in cell membranes in the human reproductive tract and adrenal gland, where it catalyzes sequential hydroxylase and lyase reactions in the biosynthesis of testosterone and other sex hormones. The approved Janssen Biotech drug abiraterone (Zytiga) and the Tokai Pharmaceuticals agent TOK-001 (Galeterone), which is in clinical trials, are prostate cancer treatments that work by inhibiting both the enzyme’s lyase activity, accounting for most of their efficacy, and its hydroxylase activity, which causes side effects.
Emily E. Scott, an associate professor of medicinal chemistry, and Natasha M. DeVore at the University of Kansas have obtained X-ray structures by getting CYP17A1 to crystallize with each of these drugs bound (Nature, DOI: 10.1038/nature10743).
The structures reveal that the drugs bind to the enzyme in a manner far different from what scientists expected. Modeling studies had suggested the drugs would orient parallel to the plane of the enzyme’s active-site heme group; instead, the drugs are nearly perpendicular to it.
Other P450 enzymes have been analyzed structurally, says cytochrome P450 specialist Paul Ortiz de Montellano of the University of California, San Francisco. But the new structures are “particularly important” in the search for prostate and breast cancer drugs with fewer side effects. For instance, they could lead to medications highly selective for CYP17A1, one of 57 similar P450 enzymes found in humans.
Scott says her group is collaborating with that of University of Kansas synthetic chemist Jeffrey Aubé “to design new compounds we hope will be more-selective drugs”—by inhibiting the enzyme’s lyase but not its hydroxylase activity, for example.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter