Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Aptamer-Guided Nanoparticles Slip into Cells
Nanomaterials: RNA molecules direct drug-loaded nanoparticles into prostate cancer cells
by Laura Cassiday
January 11, 2012

Researchers have engineered RNA molecules to guide nanoparticles into prostate cancer cells (ACS Nano, DOI: 10.1021/nn204165v). These molecules could help the particles deliver chemotherapy drugs to patients’ cancer cells, the researchers say.
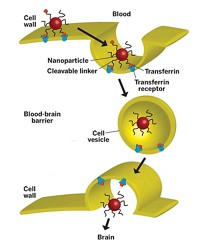
Previously, researchers have attached antibodies, other proteins, or nucleic acids to nanoparticles to target them to specific receptors on the surfaces of cancer cells. But these approaches just help the nanoparticles reach the cancer cells and don’t necessarily slip them inside to release their toxic payload, says Omid Farokhzad of Harvard Medical School. Once at the cell’s surface, the nanoparticles often must rely on simple diffusion to cross the membrane—an inefficient process, he says.
So Farokhzad, his postdoc Zeyu Xiao, and their coworkers wanted to find molecules that could help the particles take a more active route into the cells. They decided to look for folded strands of RNA called aptamers that could bind to proteins on cancer cells and trigger cellular machinery to pull in the RNA and a nanoparticle linked to it.
The researchers identified these RNA aptamers through a process called in vitro selection. First, they incubated normal human prostate cells with a pool of about 100 trillion random RNA sequences. The team discarded the aptamers that bound to these healthy cells and incubated the remaining RNAs with prostate cancer cells. To find the molecules that could make it inside, the scientists broke open the cells and collected any aptamers. The researchers then amplified these selected RNAs with polymerase chain reaction to create a new pool for selection. They repeated the entire three-step process 11 times, with each cycle weeding out inefficient aptamers and increasing the concentration of the highly efficient ones. Finally, only 68 of the original 100 trillion sequences remained; these were the aptamers that were the most efficient at getting inside prostate cancer cells.
The team sequenced these winning aptamers and further characterized the three most abundant sequences. A shortened version of one of the aptamers, known as XEO2 mini, readily crossed into a type of prostate cancer cell called androgen-independent prostate carcinoma, but couldn’t efficiently enter any of five other types of cells: normal prostate cells, another type of prostate cancer cell, or cancer cells from the breast, cervix, and lung. “This aptamer shows exquisite specificity in cell recognition and uptake,” says Farokhzad.
To test the aptamer’s ability to deliver drugs, Farokhzad and Xiao linked XEO2 mini to lipid–polymer nanoparticles that encapsulated the chemotherapy drug docetaxel. When they treated androgen-independent prostate cancer cells with the drug-loaded nanoparticles, they found that the nanoparticles attached to aptamers killed about 20% more cells than did nanoparticles lacking the aptamer. Now the researchers plan to test the aptamer-conjugated nanoparticles in animal models of cancer.
Joseph DeSimone, a chemist at the University of North Carolina, Chapel Hill, calls the technology promising. He says that the RNA selection method is powerful because it allows researchers to identify potential drug delivery aides without prior identification of proteins present on the surfaces of cancer cells.
This story was updated on Jan. 12, 2012, to correct Joseph DeSimone's field of study.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter