Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
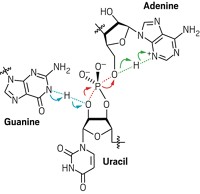
DNA
A Twist To DNA Base Pairing
Structural Biology: NMR spectroscopy proves the existence of an alternative to Watson-Crick base pairing
by Erika Gebel
February 17, 2012

DNA is one of the most intensely studied molecules, and scientists continue to uncover new layers of structural complexity. Now researchers have found direct evidence from NMR experiments that DNA bases occasionally use a little-known set of hydrogen bonds to pair up with their partners (J. Am. Chem. Soc., DOI:10.1021/ ja2117816).
In 1953, James Watson and Francis Crick described the hydrogen bonds that help DNA bases pair up. A decade later, Karst Hoogsteen recognized that bases had the potential to pair up in an alternate way: The purine bases, adenine and guanine, could flip their normal conformations and form a new set of hydrogen bonds with their partners. Most biochemists didn’t think this so-called Hoogsteen base pairing played a significant role in double helical DNA’s structure.
Then last year, Hashim Al-Hashimi of the University of Michigan, Ann Arbor, surprised biochemists by spotting Hoogsteen base pairing in a double helix (Nature, DOI: 10.1038/nature09775). The study relied on indirect nuclear magnetic resonance data from the purines’ carbon atoms, which don’t directly participate in hydrogen bonding.
To strengthen the claim, Al-Hashimi and his team developed a technique to monitor the purines’ nitrogen atoms, which take part in the bases’ hydrogen bonds. They studied short strands of DNA using a method called NMR relaxation dispersion, which can detect transient structural conformations. The researchers found that signals for certain nitrogen atoms changed over time, indicating to the researchers that some of the nitrogen atoms lost a hydrogen bond, while others gained one. The pattern matched the one that the researchers expected to occur during the transition between Watson-Crick and Hoogsteen base pairing.
While the new pairing occurred on average only 1% of the time, the team observed that certain DNA sequences increased the preference for Hoogsteen bonding. For example, adenines following cytosines were the most likely to flip and adopt the alternate bonding.
Al-Hashimi suggests that Hoogsteen base pairing could provide DNA-binding proteins with a feature for molecular recognition. “You can imagine,” he says, “that nature may have evolved systems that can recognize these structures.”


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter