Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Synthesis
Simple Probe Allows Drug Candidate Screening Without Labels
Drug Discovery: A small molecule fluoresces only when dislodged from a protein target by a drug molecule
by Erika Gebel
April 27, 2012

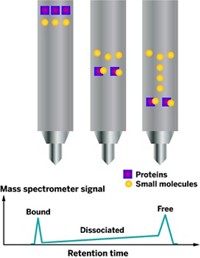
Researchers have designed a new small-molecule probe that binds to a protein and emits a fluorescent signal only when a drug molecule displaces it (J. Am. Chem. Soc., DOI: 10.1021/ ja301204z). The chemists hope that the probe will provide a simple, rapid method to screen drug candidates.
“There is a need for easy systems for screening drugs,” says Sankaran Thayumanavan of the University of Massachusetts, Amherst. Most fluorescence-based methods allow for fast screening, but they require researchers to tag the protein target or drug candidate with dyes. Tagging adds another step of planning and design to the screening process.
To skip the labeling step, Thayumanavan and his colleagues designed a probe molecule that would fluoresce only when a drug candidate bound the target.
They selected a well-characterized protein, human carbonic anhydrase I, as their initial target. The chemists’ probe used a linker containing an ester group to connect arylsulfonamide, a molecule known to bind the protein, to 7-hydroxy-4-methylcoumarin, a fluorescent dye. While attached to the linker’s ester group, 7-hydroxy-4-methylcoumarin stays dark when hit with ultraviolet light. But as soon as an enzyme called an esterase cleaves the linker’s ester and frees the dye, it fluoresces.
The researchers had previously observed that when a small molecule binds to proteins, nearby enzymes can’t react with it until a second molecule dislodges it. Based on those observations, the team thought that the probe would stay dark when bound to carbonic anhydrase. But when a drug molecule pushed it out, esterases could snip the probe’s linker to activate its dye.
To test this method, the researchers combined the probe with carbonic anhydrase and an esterase. When they excited the mixture with ultraviolet light, the solution remained dark. But when they added a compound known to bind the target, the solution started to glow, confirming that the ligands had dislodged the probe. As the chemists increased the concentration of the compound, the fluorescence intensity grew.
The researchers repeated the experiment with five compounds also known to bind carbonic anhydrase. Molecules with high affinity for the protein produced a faster increase in fluorescence than those with low affinity did. While the researchers couldn’t use the method to determine a chemical’s exact affinity for the target, they could separate weak binders from tight ones.
“I think the idea is pretty cool,” says Eric Anslyn of the University of Texas, Austin. “It’s very simple, which is always great, and seems broadly applicable.” Researchers could attach the glowing end of the probe to a drug candidate, he says, and then use it to screen for molecules that bind more tightly to a target. Anslyn hopes the researchers will next use the method on a less characterized protein, to see if it works when not all the details are known in advance. Thayumanavan has planned such experiments with protein targets that the pharmaceutical industry focuses on.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter