Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
Tool May Aid Biosynthetic Reverse Engineering
Chemical Biology: Researchers reversibly tag a protein essential for the assembly of complex molecules in biological cells
by Sarah Everts
September 19, 2012

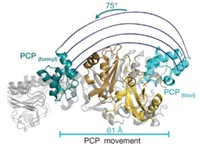
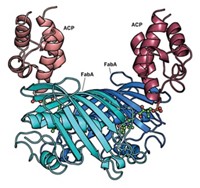
Some of nature’s most complex molecules are made by cellular factories that rely on a protein called ACP to shuttle growing molecules along biological assembly lines. Now, chemists in California are reporting the first method for reversibly labeling this protein.

The technique could help researchers learn how to successfully rewire cellular factories—such as those involved in polyketide, nonribosomal protein, and fatty acid syntheses—to allow construction of new, economically relevant products, such as drugs or biofuels, says Michael D. Burkart, a chemist at the University of California, San Diego.
Nearly 10 years ago, Burkart’s team invented a way to attach a phosphopantetheine label onto ACP’s hydroxyl “handle.” The phosphopantetheine label could be conjugated to different tags, making it easy to study various imaging, functional, and structural processes involving ACP. But the technique was limited for two reasons: The phosphopantetheine label couldn’t be removed, and ACP’s hydroxyl was often already posttranslationally modified, preventing ACP’s tagging with phosphopantetheine in the first place.
Now Burkart’s team reports that an enzyme called ACP hydrolase from the bacterium Pseudomonas aeruginosa can remove any modifications on this hydroxyl handle (Nat. Methods, DOI: 10.1038/nmeth.2175). This finding makes the ACP-labeling technique reversible, enabling study of the protein with an iterative sequence of different labels, removal of endogenous modifications, and allowing phosphopantetheine tagging for scientific studies.
Finding a way to remove modifications from ACP’s hydroxyl group took a decade to develop because the hydrolase enzyme that catalyzes removal was long mislabeled in gene databases. In addition, many hydrolases in other bacteria such as Escherichia coli were too unstable to be useful lab tools, Burkart tells C&EN.
Burkart and his team conducted proof-of-principle studies of the new technique on fatty acid synthesis in algae, they report in Nature Methods as well as in PLoS One (DOI: 10.1371/journal.pone.0042949). They hope to exploit algae fatty acid synthesis to produce biofuels in bioreactors, in collaboration with scientists at the San Diego Center for Algal Biotechnology.
One could also imagine using the new tool to understand how these biosynthetic machines work by stalling them “with a substrate analog attached to the phosphopantetheine group or by studying the interactions between an attached substrate and ACP,” comments Nenad Ban, who works with these systems at the Swiss Federal Institute of Technology in Zurich.
Mohamed Marahiel, who studies cellular factories at the Philipps University, in Marburg, Germany, calls the method an “elegant and clever” technique to reversibly label ACP proteins in fatty acid synthesis. But he’d also like to see further proof-of-principle work of the method in other biosynthetic pathways, such as polyketide synthesis and nonribosomal protein synthesis.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter