Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Pharmaceuticals
Drug Design On The Fast Track
Drug Discovery: Computer modeling approach zeroes in on compounds that can hit multiple targets and avoid side effects
by Stu Borman
December 24, 2012
Improving existing drugs and finding new ones that will treat diseases with fewer side effects could be fast-tracked by computerized mimicking and automation of drug design approaches medicinal chemists already use.
The new computational drug discovery technique, its developers say, quickly identifies and optimizes drugs that interact with multiple protein targets to combat given diseases. At the same time, the system looks to minimize or eliminate target binding that could result in serious side effects.
Its developers use it to refine existing drugs and design new ones with higher-affinity interactions and in a more automated way than has previously been possible. If the findings are confirmed, they could lead to a rethinking of the approach to drug optimization and discovery.
Several related de novo drug design methods have been proposed before, the researchers note in their paper (Nature, DOI: 10.1038/nature11691). “However, of those that have been experimentally tested, high-affinity ligands have been described only rarely, and these are all against a single molecular target objective.” The researchers’ experiments demonstrate that the new technique design compounds with high affinity for selected multiple targets.
The technique was devised by medical informatics specialist Andrew L. Hopkins of the University of Dundee, in Scotland; pharmacologist Bryan L. Roth of the University of North Carolina School of Medicine; and coworkers. Hopkins has started a company, Ex Scientia Ltd., in Dundee, to commercialize the technology.
The approach is an automated, computational version of techniques medicinal chemists use to optimize structure-activity relationships (SARs) of drug-lead compounds, but at multiple targets. It uses information in databases on drug-target interactions to tweak compound structures in an iterative manner to enhance activity and reduce side effects until these figures of merit reach preset goals.
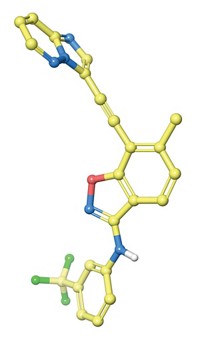
For example, the team took the existing acetylcholinesterase inhibitor donepezil and modified it computationally to hit multiple targets completely different from its original one. To do this, they used Bayesian analysis—an adaptive technique for making inferences from limited data—to score library compounds on their likelihood of penetrating the central nervous system and for good absorption, distribution, metabolism, and excretion properties. An algorithm filtered the best candidates for novelty, adherence to the “Rule of Five” structural and physical properties shared by most successful drugs, and ease of chemical synthesis. They repeated the optimization process numerous times until they identified an agent that adopted desired multitarget activities and avoided off-target activities.
Computational drug design specialist Brian Shoichet of the University of California, San Francisco, comments that the researchers’ major innovations are that “they computationally represent SARs, provide objective goals for their drug designs to meet, and draw on SAR data for about 800 drug targets. It would normally be difficult for medicinal chemists to keep track of SARs for that many targets.”
It remains to be seen whether medicinal chemists find the new approach to be a practical strategy they wish to adopt, Shoichet adds. Nevertheless, “it will resonate with medicinal chemists because it automates the type of SAR analysis they are already familiar with,” he says.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter