Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Physical Chemistry
Simulation Breaks Time Barrier
Computational Chemistry: Made with surplus computing power, prediction of how receptor works is lengthiest by far
by Stu Borman
December 23, 2013
| A version of this story appeared in
Volume 91, Issue 51

Given that G protein-coupled receptors (GPCRs) regulate many biological processes and are targets of about one-third of marketed drugs, scientists are eager to learn as much as possible about how they function. A new theoretical study carried out in the “cloud”—on Internet computer servers—reveals new evidence about how a key GPCR likely works, information previously inaccessible with dedicated lab computers. The feat confirms that Internet computing could contribute to future insights into biological problems.
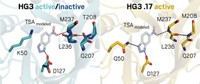
The study used Exacycle, a research program that permits scientists to run parallel computations on Google’s servers when idle. A Stanford University-Google group used Exacycle to carry out a molecular dynamics simulation of activation and deactivation of the GPCR β2-adrenergic receptor, a protein that plays a role in diabetes, obesity, and asthma. Molecular dynamics predicts physical movements of atoms in a protein or other molecule as it functions.
The simulation watched the receptor’s actions for two milliseconds—about an order of magnitude longer than ever before. The previous record, several hundred microseconds, was achieved on a computer designed specifically for molecular dynamics.
The study was carried out by Kai J. Kohlhoff of Stanford and Google; Diwakar Shukla, Morgan Lawrenz, Russ B. Altman, and Vijay S. Pande of Stanford; and coworkers (Nat. Chem. 2013, DOI: 10.1038/nchem.1821). They performed their molecular dynamics computations on Exacycle and then used a technique called Markov state modeling to string the computed pieces together into a simulation of β2-adrenergic receptor activation and deactivation. The simulation agrees with previous computational and experimental results and provides new information about how signaling molecules affect receptor function.
“Even setting the interesting biological results aside, a simulation this long represents an important methodological milestone,” comments computational chemist Jacob Durrant of the University of California, San Diego. The technique could lead to “more accurate computer-aided drug design methods for predicting small-molecule binding, thereby improving the hit rates of subsequent validation experiments and potentially accelerating the drug discovery process.”
Molecular dynamics expert Ursula Röthlisberger of the Swiss Federal Institute of Technology, Lausanne, notes that the approach currently requires preexisting structural information about the target, and “it will really be interesting to see if it can be extended to cases where the active form has not been crystallized.” Nevertheless, she says, the study shows that the cloud can be used “to tackle some of the hardest computing problems we have at the moment.”


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter