Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Biological Chemistry
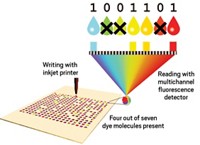
Scientists Look To DNA For Long-Term Data Storage
ACS Meeting News: Encoding DNA with digital information and encasing it in silica nanoparticles could save documents and data for future generations
by Stephen K. Ritter
September 7, 2015
| A version of this story appeared in
Volume 93, Issue 35

DNA is known for being a storehouse of genetic information. Just one gram of the genetic material can theoretically hold some 300,000 terabytes of information, far exceeding the roughly 5 terabytes found on the best computer hard drives. And as a result of finding intact DNA in fossilized bones, scientists know that DNA can remain stable for hundreds of thousands of years, whereas hard drives might last only 50 years.
Those attributes have made synthetic DNA a desirable medium for long-term digital information storage to preserve historical documents and important data for future generations. One limitation, however, has been a practical way to encode information into DNA and then later retrieve it on demand without errors caused by degradation of the DNA or mistakes in sequencing.
Robert N. Grass of the Swiss Federal Institute of Technology, Zurich, and his colleagues have been working to solve those issues and are beginning to achieve success by storing encoded DNA within silica nanoparticles. Grass reported his team’s latest results during a symposium organized by the Division of Industrial & Engineering Chemistry at the American Chemical Society national meeting in Boston last month.

When DNA’s structure was solved some 50 years ago, scientists found that the coding language of nature is similar to the binary language we use in computers, Grass explained. On a hard drive, a series of 0s and 1s representing data are stored on a magnetic material. In DNA, the four nucleotides that make up the biopolymer—adenine, cytosine, thymine, and guanine—go by the letters A, C, T, and G. Scientists can use those letters to represent data by synthesizing DNA with a coded sequence of nucleotides.
DNA data storage is not new; encoding digital information into DNA was first demonstrated in 1988. Since then, scientists have achieved moderate success as sequencing technology has improved.
For example, in 2012 Sriram Kosuri of Harvard Medical School and the Wyss Institute for Biologically Inspired Engineering and coworkers showed that the text of a short synthetic biology book could be stored and retrieved using commercially available DNA synthesis and sequencing methods (C&EN, Aug. 20, 2012, page 29). And late last year, Kosuri teamed up with the band OK Go to begin encoding their album “Hungry Ghosts” on DNA. If the project comes to fruition, fans could buy a vial with a few nanograms of DNA dissolved in a drop of water that contains some 100,000 copies of the album.
In Boston, Grass provided a simplified description of his team’s way of encoding DNA. For example, the text string “C&EN” could be represented as C = CTG, & = ACT, E = ATC, and N = GGA, which overall would be CTGACTATCGGA. But the coding is not as straightforward as that, he said.
Just in case the DNA degrades or some of the data are corrupted, Grass and his team developed an error-correction code that they build into DNA sequences. This extra code requires additional nucleotides be added to the sequence.
Error-correction codes, first developed more than 50 years ago as an elegant solution to prevent errors in early satellite communication, use polynomial equations to set up redundant pieces of information. For DNA storage, even if one section of the DNA is not readable, or if an error occurs in sequencing the DNA, reading the full DNA sequence with the redundancies still allows retrieval of all the information error-free. In the ETH Zurich approach, this means every two letters of a text or data file end up being mapped to nine nucleotides.
DNA is a good long-term storage tool because people likely will always have the need for sequencing DNA and the technologies to do so, Grass added. But his team still needed to find a better way to store DNA for centuries, or perhaps longer. Inspired by retrievable DNA found in fossils, Grass and his coworkers thought they could make “synthetic fossils” by encapsulating DNA in silica nanoparticles.
Once the ETH Zurich researchers sequence the encoded DNA, they use amino linking groups to bind DNA strands to the surface of silica nanoparticles. Using a sol-gel process, they coat the DNA with additional silica—effectively adding a layer of protective glass. When the DNA needs to be retrieved, the researchers dissolve the silica using a weak, buffered solution of hydrofluoric acid. To access the information, they then sequence the DNA (Angew. Chem. Int. Ed. 2015, DOI: 10.1002/anie.201411378).
To test their storage system, Grass and his team encoded DNA with text from the Swiss Federal Charter of 1291 and an English translation of Archimedes’ “The Method of Mechanical Theorems.” Rather than saving the information on one long DNA strand, they synthesized some 5,000 DNA strands each 158 nucleotides long.
The team heated the DNA-loaded nanoparticles at 70 °C for a week to simulate aging equivalent to 2,000 years of storage at room temperature. They then recovered the DNA and decoded the digital information without any errors in the text.
For now, the method is limited to reading the whole string of data—the researchers can’t point to a specific place in a passage of text and read it. So the ETH Zurich team is working toward developing a way to label pieces of information to make a document searchable.
According to Kosuri, who is now at the University of California, Los Angeles, the ETH Zurich researchers have made important advances in determining DNA degradation rates, provided an easy method for DNA encapsulation and recovery, and developed perhaps the most reasonable error-correcting schemes so far. “I think this is a well-thought-out and thorough examination of where things stand in the DNA encoding space,” Kosuri told C&EN.
Grass admitted the data storage method might be useful only for saving historical documents and large permanent databases. But his group is looking to apply the silica technology elsewhere. For example, the researchers have used silica nanoparticles for preserving RNA for biomedical research. RNA is usually stored in freezers and shipped packed in dry ice. But with silica protection, or with protection in other types of matrix materials, the need for cold storage could become obsolete, he believes.
“DNA storage is a very important and timely topic, given the trends in reducing synthesis and sequencing costs and the emergence of the big data era,” commented electrical and computer engineering professor Olgica Milenkovic of the University of Illinois, Urbana-Campaign. In fact, Milenkovic noted that two other gatherings featuring research on bioinspired computing and DNA data storage took place in the Boston area at the same time as the ACS meeting: the DNA21 conference on DNA computing and molecular programming at Harvard University and a Massachusetts Institute of Technology workshop on distributed algorithms inspired by biology.
“Grass’s work takes a fresh new look at the problem from the perspective of determining the ‘best’ storage conditions for this new nonvolatile medium,” she said.


Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter