Advertisement
Grab your lab coat. Let's get started
Welcome!
Welcome!
Create an account below to get 6 C&EN articles per month, receive newsletters and more - all free.
It seems this is your first time logging in online. Please enter the following information to continue.
As an ACS member you automatically get access to this site. All we need is few more details to create your reading experience.
Not you? Sign in with a different account.
Not you? Sign in with a different account.
ERROR 1
ERROR 1
ERROR 2
ERROR 2
ERROR 2
ERROR 2
ERROR 2
Password and Confirm password must match.
If you have an ACS member number, please enter it here so we can link this account to your membership. (optional)
ERROR 2
ACS values your privacy. By submitting your information, you are gaining access to C&EN and subscribing to our weekly newsletter. We use the information you provide to make your reading experience better, and we will never sell your data to third party members.
Drug Development
Harnessing The Hordes In The Microbiome
As researchers learn more about the connection between our bacteria and our health, companies are trying to develop therapies that exploit it
by Lisa M. Jarvis
September 28, 2015
| A version of this story appeared in
Volume 93, Issue 38

We humans are born alone. But that solitude is fleeting. Even as we emerge from the womb, we’ve started to accumulate the first few hundred of our lifelong companions—the rich population of microbes that live in and on us. By the time we greet our kindergarten teacher, we are outnumbered: The microbial cells we carry far exceed our own.
These bugs aren’t freeloaders. We maintain a symbiosis with our microbiome that helps keep us healthy. Its job includes breaking down foods our bodies are otherwise unable to digest, metabolizing nutrients into needed vitamins, helping to regulate glucose levels, and sending signals to our immune system.
“We’ve got a hundred trillion bacteria in and on our bodies that have coevolved with us from the dawn of humanity,” notes Peter DiLaura, chief executive officer of the microbiome-focused biotech firm Second Genome. “In many ways, we have colonized the bacteria as opposed to the other way around.”
When the balance in that vibrant community is disrupted—known as “dysbiosis”—the health of its host can suffer. Since the 2008 launch of the Human Microbiome Project, the National Institutes of Health’s effort to map what lives in us and where, the microbiome field has seen an explosion of publications drawing connections between our bacteria and our health.
In many cases, the findings are merely correlations. Researchers still need to prove that an imbalance in the microbial community is a cause or contributor, rather than the consequence, of disease.
But the evidence that our commensal bacteria—our everyday bugs—play a role in disease is compelling enough to lure researchers and investors into developing therapies based on or targeting them. The past five years have seen a proliferation of companies with multiple approaches to leveraging the interaction between our bugs and our bodies.
Efforts include whole-cloth microbiome replacements, rationally designed microbial cocktails, and small molecules that disrupt a specific interaction. Still other groups are considering the complex natural products made by our microbiota.
Overall, some 60 companies are publicly working on microbiome-related projects, with many more under the radar, notes Dirk Gevers, head of Johnson & Johnson’s Janssen Microbiome Institute. And beyond the flourishing biotech community, big pharma firms are dipping their toes into the water. J&J has made the most significant investment, but nearly every major firm has some effort—either partnerships or internal activities—to develop microbiome-modulating therapeutics.
AN INFLECTION POINT

Several technological and clinical advances have converged to produce the current microbiome mania. The most critical development has been the ability to use gene sequencing to quickly identify the microbes camping out in the human body and sometimes deduce their function.
“To understand the microbiome, we have to look at literally thousands of different species of bacteria all at once,” says Roger Pomerantz, CEO of Seres Therapeutics. Without the precipitous drop in the cost and time it takes to capture and analyze gargantuan quantities of genetic data, he adds, “the microbiome revolution would not be here.”
The new sequencing capabilities have enabled projects that not long ago would have been inconceivable. For example, four-year-old Second Genome recently did an experiment that required it to sequence and analyze several thousand biological samples. Whereas a decade ago it would have taken a year to sequence just one genome, the company’s researchers wrapped up the project in just a month.
Beyond sequencing a massive number of genomes, Second Genome’s project hinged on the ability to analyze and cheaply store data in the cloud. “If I had to do this just five years ago, I would have had to buy lots of servers and build all of this infrastructure, which still wouldn’t be as good as what I can immediately pull up on demand,” DiLaura says.
The technological advances dovetail with compelling clinical evidence that our microbial companions play a role in our health. Before companies with the label “microbiome” even existed, infectious disease doctors knew the power of altering our bacterial makeup: They learned to cure a dangerous gut infection by replacing a sick microbial community with a healthy one.
When a hospital patient takes broad-spectrum antibiotics to battle an infection, one pernicious bug called Clostridium difficile is able to evade death by forming protective spores. After the other bacteria are licked, C. difficile roars back and dominates the gut, causing serious diarrhea and abdominal pain.
Desperate to treat people infected by this potentially deadly bug, a small number of doctors in recent years turned to a method that sounds odious: fecal microbiota transplantation, or repopulating the bacteria in a patient’s colon using a stool sample from a healthy volunteer. Fecal transplants have been shown to cure up to 90% of the very sickest C. difficile patients. Not only that, but the response is durable. For most patients the infection doesn’t come back.
THE FIRST WAVE

Success treating C. difficile with fecal transplants demonstrated the therapeutic potential of harnessing the microbiome. Given the concentration of bacteria in the gut, it’s not surprising that the first microbiome-based therapies in development are focused on infections or disorders that affect our gastrointestinal tract. They include C. difficile infection and inflammatory bowel diseases like ulcerative colitis and Crohn’s disease.
Scientists have long known that gut microbes are critical for certain human functions. The trillions of microbes churning in our gut consume oxygen, secrete peptides and enzymes, help break down polysaccharides, and play a role in synthesizing amino acids and short-chain fatty acids. “It’s a massive bioreactor,” Second Genome’s DiLaura quips.
The most obvious way to broach the gut microbiome is by riffing off the success of the fecal transplant. Several companies are developing pills that simply encapsulate fecal samples from healthy people. Seres Therapeutics has fine-tuned the approach with a pill containing spores of beneficial bacteria derived from stool samples. SER-109 is in Phase II trials to prevent C. difficile from recurring.
Moving beyond the first generation of therapies, Vedanta Biosciences is developing bacterial cocktails on the basis of biological insights by the company’s academic founders into the interplay between our microbiota and our immune system. Vedanta started with the observation that the gut microbiota is critical for the development of Tregs, immune regulatory cells that turn down the inflammatory response and also play a role in inflammatory bowel disease.
Kenya Honda of Japan’s Keio University developed a way to screen for bacteria that prompt the immune system to turn up the activity of Tregs, ultimately creating the cocktail that became Vedanta’s lead therapy, VE202. He later determined the relationship between the beneficial bacteria and their host: Clostridia, a class of commensal microbes found in abundance in our gut, produces the short-chain fatty acid, butyrate, that prompts Tregs into action.
“By drilling down to the mechanism of action, we could systematically screen different microbes for their effects on the host immune system,” says Vedanta CEO Bernat Olle. On the basis of that screen, 17 strains of Clostridia that pushed Tregs into action were combined to become VE202.
J&J bought the rights to develop VE202 as a treatment for inflammatory bowel disease earlier this year. Vedanta meanwhile is building a pipeline of other therapies that modulate the relationship between the microbiome and the host immune system.
One advantage of treatments such as those being developed by Vedanta and Seres is the possibility of an easy—or at least fast—path from concept to clinic. Because their therapies are simply strains of bacteria that already populate our bodies, concerns about safety are minimal. “You’re dealing with human bacteria that are commensals,” Seres’s Pomerantz says. “They’ve evolved with us for thousands of years.”
Seres didn’t have to conduct a traditional Phase Ia trial to study the effect of different doses of its lead product, SER-109. “This is a living drug that goes into your colon, and the spores turn back into bacteria,” Pomerantz says. “In two days it’s a thousand fold of what you put in.”
MAKING CONNECTIONS

Underlying all the enthusiasm is the proviso that most of the connections between the microbiome and human health are just that—connections. Although dysbiosis has been correlated with many diseases, researchers still need to prove it is causing or contributing to those diseases and not the consequence of them.
“There’s this chicken-and-egg problem that’s going to be really important to solve when it comes to thinking about whether microbes are good therapeutic targets,” says Harvard University chemist Emily Balskus.
Chemists like Balskus are confident they can help determine what’s the chicken and what’s the egg. To date, microbiome research has focused on figuring out which microbes are populating an organ rather than understanding what they do. Now, academic and industrial scientists alike are drilling down into the mechanistic details of the microbiome-host interaction with the goal of developing small molecules to modulate that relationship. But not until the molecules work will they know for sure what’s driving disease.
University of North Carolina chemist Matt Redinbo was among the first to show it is possible to use a small molecule to safely target gut bacteria, a critical discovery for those contemplating modulating the microbiome.
Redinbo’s breakthrough began out of curiosity: Why did the colon cancer drug irinotecan cause severe diarrhea? Other researchers had shown that irinotecan, a prodrug with a dipiperidino group that is clipped off to form an active metabolite, is inactivated in the liver by the addition of glucuronic acid.
Redinbo found that when the inactive compound reaches the intestine, otherwise beneficial gut microbes scavenge off the added sugar, turning the drug’s cell-damaging properties back on again. The reactivated drug kills the epithelial cells that line the gastrointestinal tract, causing diarrhea so severe that some colon cancer patients stop taking it all together.
Redinbo’s lab set out to block the bacterial enzymes responsible for scavenging the sugar group. The task was not simple. To work as drugs, compounds would need to bind to the bacterial β-glucuronidase, but not the human analog, and also avoid killing the otherwise beneficial bacteria.
Then Redinbo, by his own admission, got “superhuman lucky.” A high-throughput screen yielded β-glucuronidase inhibitors with up to 1,000-fold selectivity for the bacteria. After solving the crystal structure of one molecule bound to the target enzyme, his lab found a 20-amino acid loop covering the active site that is unique to the bacterial enzyme. The compounds were binding to that loop and thus not hitting the analogous enzyme in humans.
Redinbo started a company called Symberix to develop molecules to alleviate the diarrhea that limits the dosing of many cancer drugs. The company has government funding and is optimizing hits that came out of his lab’s screen.
Other labs are slowly picking apart microbial biochemistry that could be implicated in human disease. Balskus’s lab at Harvard is one of several trying to decipher the mechanism by which microbes turn nutrients or other compounds into deleterious metabolites. Such work could open the door to small molecules or other treatments to disrupt those pathways.
Balskus has focused on elucidating the process by which choline, a nutrient found in eggs and meat, is metabolized by bacteria. She got interested in choline metabolism after the Cleveland Clinic’s Stanley Hazen linked trimethylamine-N-oxide (TMAO), which is generated when gut microbes metabolize choline, to heart disease.
Intrigued that heart disease could be driven by our microbiota as well as our genetics, Balskus set out to determine which gut bacterial enzymes were mediating the choline fermentation. Her group discovered that choline TMA-lyase, a new member of the glycyl radical enzyme family, cleaves a C–N bond in choline to produce the TMAO precursor trimethylamine (TMA) (shown).
Her lab continues to explore how choline TMA-lysase works and what members of the gut microbiota actively use it. Balskus also has started to develop small molecules that modulate choline degradation and block TMA production, but she stresses that the work is preliminary.
THE MICROBIOME’S MOLECULES
Advertisement
Whereas Balskus and Redinbo are exploring mechanistic details of known microbe-human relationships, University of California, San Francisco, chemist Michael Fischbach thinks it’s worth considering the rich tapestry of molecules being churned out by our microbiota. It turns out that our commensal bacteria make complex natural products that could be helping—or hurting—us.
Fischbach came to the microbiome field by accident. His group was focused on mining the genomes of soil bacteria—the source of most of our antibiotics—for natural products. But around 2010, Fischbach used his computational algorithm, which allows him to mine sequencing data for gene clusters that represent biosynthetic processes, to search every genome in the NIH database.
The results surprised him. “We learned that some of the most interesting genes we were finding were not in the organisms we were used to looking at,” Fischbach recalls, referring to bacteria that live in soil or water. “They were in the recently deposited genome sequences of gut, skin, and oral bacteria.”
He was searching at a time when results from the Human Microbiome Project were being made public, and it made him ponder how to explore the molecules made by our microbiota. Although many researchers have over the years considered the health impact of the short-chain fatty acids it makes, Fischbach was convinced that wasn’t the whole story. They will be “one of what will be many dozens” of small molecules made by our microbiota that play a role in human health, he predicts.
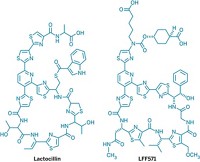
Fischbach’s lab has already produced one example of the potential pharmacopeia emerging from our microbiome. Last year, his group identified lactocillin, a thiopeptide antibiotic produced by vaginal bacteria.
Fischbach concedes that bacteria living in humans are likely less prolific producers of natural products than ones in the soil or sea. And indeed, lactocillin is one of only a handful of microbiome-made natural products to be well characterized to date. “However, that doesn’t mean the smaller number is going to be less important,” he says. “I think the molecules are worth finding.”
SHIFTING SIGNALS
While academic chemists strive to understand the role of microbial chemistry in human health, companies are already working on small molecules that modulate specific microbe-human interactions.
The most advanced player in the small-molecule space is South San Francisco-based Second Genome, which wants to generate a pipeline of products built on a precise understanding of the gut microbe-human interaction. Whereas most researchers studying inflammatory diseases such as inflammatory bowel syndrome gauge gut microbe populations by stool samples alone, Second Genome is also interested in the community present in mucosal biopsies taken during a colonoscopy.
“Fecal samples are useful,” DiLaura says. “You just have to recognize they might not be the full complement of the microbiota at the site of disease.”
Moreover, he adds, taking a sample directly where inflammation is occurring allows the company to look not only at differences in bacterial expression between healthy and diseased tissue but consider changes in human gene expression as well.
Like everyone else, Second Genome starts a drug discovery campaign by taking a census of the bacteria living in patient tissue and stool samples and compares them with samples taken from healthy people. “But the thing that’s most relevant in a drug discovery paradigm is to then get to the biological function,” DiLaura says.
The company has technology to interpret the transcriptional output of the microbiome, allowing it to generate hypotheses about what molecules the microbiome is making and how the host is responding.
The next step for Second Genome is to test those hypotheses. The company also has in vitro assay technology that allows it to screen molecules that might modulate the microbiome-host interaction.
The most advanced compound to emerge from that process is SGM-1019, which blocks a target in the colon, thereby turning down the effect of a microbiome-made ligand implicated in inflammatory bowel disease. Second Genome began Phase I studies of SGM-1019 earlier this year, and DiLaura expects to move it into Phase II trials next year.
A PATH FORWARD
As the first therapies based on or targeted against the microbiome reach the clinic, researchers acknowledge they have plenty to learn. They need better ways to understand when to intervene with a therapeutic, Vedanta’s Olle points out, and to understand the long-term implications of altering our microbiome.
That will mean tracking the microbiome to understand how it evolves from birth to adulthood. Scientists want to be able to identify signs of dysbiosis and learn why the balance is faltering. “One of the biggest challenges is to make sure what you’re seeing is linked to disease and not some environmental or other factor,” Janssen’s Gevers says. “A key piece will be developing the right biomarkers” to assess how and why perturbations to the microbial community are occurring.
Other researchers stress that they need to better understand the microbial population at the site of disease, rather than gauging dysbiosis from a stool sample. The ability to map the microbial variation throughout the gut, and even take samples over time, also would help, notes Jim Brown, who leads GlaxoSmithKline’s microbiome matrix team.
Every player in the field acknowledges that sorting through these unknowns will help the field move beyond the obvious targets—C. difficile and inflammatory bowel diseases—and into more complex diseases such as diabetes or obesity.
Researchers stress that modulating the microbiome isn’t a cure-all. Rather, they suggest that ignoring our bacterial companions leaves out one of many possible keys to unlocking a disease.
“Modern drug discovery has been pretty narcissistic in thinking about host biology,” Second Genome’s DiLaura says. “We have this whole commensal organ sitting on top of host pathways. When you integrate what the microbiome and host are doing together, you have a much more complete picture of disease and wellness.”
SYNTHETIC BIOLOGY
Coaxing Our Companions Into Acting Like Drugs
Synlogic is pairing knowledge of the microbiome with synthetic biology to treat rare metabolic disorders
The field of microbiome-targeted therapeutics has blossomed in the past five years as evidence mounts that the bugs that live in and on us play a critical role in our health. Whereas most companies are trying to change the composition of the microbial community or disrupt signals the bugs are sending us, Cambridge, Mass.-based Synlogic is using synthetic biology to coax our gut bacteria into sensing and fixing disease.
For decades, drug companies have engineered bacteria to produce proteins or enzymes of interest. Synlogic’s synthetic biology approach goes further. Starting with microbiome-derived bacteria, the biotech firm inserts a circuit into the genome that performs an entire therapeutic task.
“Several genes in that circuit perform metabolic conversions that prompt bacteria to take a toxic metabolite and build something that is good for you,” explains Synlogic CEO Jose-Carlos (JC) Gutiérrez-Ramos.
Synlogic is initially focused on teaching our gut bacteria to treat rare metabolic diseases. One of its first programs addresses urea cycle disorders, in which a genetic mutation renders patients unable to convert nitrogen into urea. The result is a toxic build-up of ammonia in their blood. Synlogic engineers have programmed bacteria to convert ammonia to arginine or citrulline, amino acids that people with the disorders lack.
The circuit inserted into the bacteria also allows the company to control when and for how long the engineered metabolic process is active. In the case of the urea cycle disorder program, the bacteria are noncolonizing and active for less than a day.
Synlogic expects to have initial conversations with the Food & Drug Administration later this year about the urea cycle disorder program and a second treatment in its pipeline for a rare metabolic disorder called phenylketonuria. The biotech firm, which secured its first round of financing just over a year ago, hopes to start its first clinical trial at the end of 2016.



Join the conversation
Contact the reporter
Submit a Letter to the Editor for publication
Engage with us on Twitter